Introduction
Cockroach allergies are type I hypersensitivities
mediated by immunoglobulin E (IgE) and have been recognized as such
since 1964 (1). Cockroach
allergies are associated with the development of asthma and have
been shown to put patients with asthma at risk of requiring
emergency medical aid. This is a particular issue for asthmatic
children in less affluent urban households, where cockroach
infestations are more prevalent (2). Several proteins from cockroaches are
potent environmental aeroallergens. Skin prick tests in China
revealed that 25.7% of patients with allergies reacted positively
to the American cockroach Periplaneta americana (Per a
allergens) and 18.7% exhibited positive responses to the German
cockroach Blattella germanica (Bla g allergens) (3). Among the indoor cockroaches, the
American cockroach is the dominant species and is responsible for a
large proportion of the cockroach-induced allergic reactions
worldwide. Seven allergens from Periplaneta americana have
been characterized: Per a 1 (4),
Per a 2 (5), Per a 4 (6), Per a 6 (7), Per a 7 (8), Per a 9 (9) and Per a 10 (10). Per a 6 is a troponin C protein
identified in the American cockroach. It was found that native Per
a 6-bound IgE was present in the sera of 17% of Thai individuals
with cockroach allergies, indicating that Per a 6 is a minor
cockroach allergen (11).
To date, the use of cockroach immunotherapy has been
rare and there are few reports on its efficacy (12,13).
Elucidation of the B- and T-cell epitopes of allergens may enhance
the understanding of the structure-function relationship and
predict the basis of cross-reactivity. Cross-reactive epitopes may
be useful in reducing the number of allergens without compromising
the efficacy of therapy (14).
B-cell epitopes can be applied in the diagnosis, therapy and
development of effective vaccines for immunotherapy. Numerous
techniques can be utilized in the identification of B-cell
epitopes; in particular, computational tools have been suggested to
represent a promising and rapid approach. Following identification,
the allergenicity of an allergen can be reduced by modifying the
predicted B-cell epitopes (15).
In the last decade, T-cell epitopes were successfully identified
based on computer simulation. To stimulate T-lymphocyte responses,
extracellular peptides bind to major histocompatibility complex
(MHC) class II molecules (16).
Therefore, T-cell epitopes were indirectly predicted by the
identification of MHC-binding molecules (17). In the present study, the B- and
T-cell epitopes of the Per a 6 allergen and its analog in the
German cockroach, the Bla g 6.01 allergen, were identified by an
in silico approach. The results indicated the potential
utility of the B- and T-cell epitopes in a peptide-based vaccine
design for cockroach allergy.
Materials and methods
Sequence retrieval
The primary sequence of the Per a 6 allergen was
acquired from the nucleotide database of the National Center for
Biotechnology Information (NCBI; http://www.ncbi.nlm.nih.gov/) with the accession
number AY792950.1. The Basic Local Alignment Search Tool (BLAST;
http://blast.ncbi.nlm.nih.gov/Blast.cgi) was used to
obtain the homologous sequences.
Physiochemical analysis
Physiochemical analysis of the Per a 6 allergen and
its homologous sequences, including analysis of the molecular
weight, theoretical isoelectric point (pI), amino acid composition,
instability index, aliphatic index and grand average of
hydropathicity (GRAVY), was performed using the ProtParam tool
(http://web.expasy.org/protparam/).
Phylogenetic analysis
The complete amino acid sequences of the Per a 6
allergen and its homologs were aligned using Clustalx 2.1 (18). The phylogenetic tree was obtained
using the neighbor-joining (NJ) method on the basis of the
Jones-Taylor-Thornton amino acid sequence distance implemented in
the Molecular Evolutionary Genetics Analysis 5 software (19). The reliability was evaluated by the
bootstrap method with 1,000 replications.
In silico prediction of B-cell
epitopes
In the Protean™ system (DNAStar, Inc., Madison, WI,
USA), four properties (hydrophilicity, flexibility, accessibility
and antigenicity) of the amino acid sequences were selected as
parameters for the prediction of epitopes. The hydrophilicity
prediction was performed by the methods of Hopp and Woods (20) and Kyte and Doolittle (21). Flexibility was analyzed by the
strategies of Karplus and Schulz (22), while the surface accessibility and
antigenicity were analyzed by the strategies of Emini et al
(23) and Jameson and Wolf
(24). The peptide regions with
good hydrophilicity, high flexibility, surface accessibility and a
high antigenic index were selected as candidate epitopes for
further investigation. In order to enhance the veracity of
prediction towards the B-cell epitopes, the amino acid sequences
were analyzed by the Protean system.
The Bioinformatics Predicted Antigenic Peptides
(BPAP) system (http://imed.med.ucm.es/Tools/antigenic.pl) and
BepiPred 1.0 server (http://www.cbs.dtu.dk/services/BepiPred/) were
additionally used to predict B-cell epitopes. The BPAP system
follows the prediction method by Kolaskar and Tongaonkar (25), while the BepiPred 1.0 server
follows a combination of a hidden Markov model and a propensity
scale method (26). The two
servers only require the amino acid sequence and provide more
straightforward results, which are combined with the
physicochemical properties of the amino acids, including
hydrophilicity, flexibility, accessibility, turns and an exposed
surface (27). The ultimate
consensus epitope results were obtained by combining the results of
the three immunoinformatics tools with the previously published
method (28). If all three methods
concluded that the sequence was not an epitope, the consensus
result was 0% epitope. However, if only one or none of the methods
predicted that the sequence was not an epitope, the consensus
result was 67% or 100% epitope, respectively. The regions whose
consensus epitope result was 67 or 100% were selected as the final
potential epitope regions.
In silico prediction of T-cell
epitopes
T-cell epitopes are principally predicted indirectly
by identifying the binding of peptide fragments to the MHC
complexes. However, the binding grooves of MCH-II molecules are
open at both ends, allowing various lengths of peptides to bind,
and the same MHC molecule can accommodate various different binding
sequences. These two properties make the development of accurate
predictive algorithms for MHC-class II binding complicated. It has
been reported that a sequence nine amino acids in length (9-mer),
known as a core peptide, is essential for MHC-class II binding
(29). For human leukocyte antigen
(HLA)-DR-based T-cell epitope prediction, the artificial neural
network-based alignment (NN-align) method NetMHCIIpan-2.0
(http://www.cbs.dtu.dk/services/NetMHCIIpan/) was
applied. NetMHCIIpan-2.0 provides predictions only for a selection
of the most commonly found HLA-DR alleles. For HLA-DQ alleles,
NetMHCII-2.2 (http://www.cbs.dtu.dk/services/NetMHCII/) was used
(30). Although the predicted
results provided limited binding-affinity data, it was recently
reported to provide the best performance in predicting this locus
(31). For the T-cell prediction
process, the peptides predicted to be significant binders by at
least one allele were examined. The binding significance of each
peptide to the given MHC molecule was based on the estimated
strength of binding exhibited by a predicted nested core peptide at
a set threshold level. Based on the estimated strength of binding,
the peptides were characterized as significant (weak or strong) or
non-significant. Each predicted core 9-mer sequence represented the
predicted putative T-cell epitope within each peptide examined
(32).
Results
Physiochemical analysis and phylogenetic
analysis
The amino acid sequence of the Per a 6 allergen was
obtained from the nucleotide database of the NCBI. The primary
structure contained 151 amino acids. A search with BLAST obtained
16 homologous sequences of the allergen. The accession numbers of
these homologs were DQ279092.1 (Blattella germanica allergen
Bla g 6.01), DQ279093.1 (Blattella germanica allergen Bla g
6.02), XM_001987063 (Drosophila grimshawi GH21731),
XM_002049988 (Drosophila virilis troponinC41C),
XM_002106068.1 (Drosophila simulans GD16679), XM_002086000
(Drosophila yakuba GE11315), GQ396705.1 (Bombyx mori
troponin CIII), XM_001688761.1 (Anopheles gambiae str.
PEST), AY075534.1 (Drosophila melanogaster RH0783),
NM_078895.4 (Drosophila melanogaster troponin C), X76043.1
(Drosophila melanogaster troponin C), XM_002063468.1
(Drosophila willistoni GK21371), AB180453.1 (Plutella
xylostella troponin C), XM_001867954.1 (Culex
quinquefasciatus troponin C), AF432912.1 (Solenopsis
invicta troponin C), and NM_001205104 (Acyrthosiphon
pisum troponin C-like). Table
I shows the details of the physiochemical analysis of these
homologs. All homologs were stable (instability index ranging
between 24.97 and 38.57) with theoretical pIs ranging between 3.83
and 4.05. The GRAVY values were found to be negative (−0.457 to
−0.220) for complete sequences.
 | Table IPhysiochemical properties of the Per
a 6 allergen and its homologous sequences. |
Table I
Physiochemical properties of the Per
a 6 allergen and its homologous sequences.
| Accession no. | MW | NCR (n) | PCR (n) | TP | II | AI | G |
|---|
| AY792950.1 | 17130.9 | 44 | 11 | 3.84 | 24.97 | 87.22 | −0.377 |
| DQ279092.1 | 17216.2 | 43 | 13 | 3.96 | 35.44 | 86.56 | −0.388 |
| DQ279093.1 | 17095.1 | 43 | 13 | 3.96 | 31.48 | 87.22 | −0.269 |
| XM_001987063 | 16989.7 | 40 | 10 | 3.84 | 32.11 | 86.05 | −0.324 |
| XM_002049988 | 17557.4 | 40 | 10 | 3.84 | 29.52 | 81.58 | −0.317 |
| XM_002106068.1 | 17155.9 | 40 | 10 | 3.83 | 31.63 | 83.05 | −0.325 |
| XM_002086000 | 16937.6 | 40 | 10 | 3.83 | 29.02 | 84.14 | −0.336 |
| GQ396705.1 | 17275.3 | 42 | 13 | 4.03 | 34.45 | 86.01 | −0.291 |
| XM_001688761.1 | 17609.5 | 42 | 12 | 3.90 | 27.87 | 85.00 | −0.384 |
| AY075534.1 | 17155.9 | 40 | 10 | 3.83 | 31.63 | 83.05 | −0.325 |
| NM_078895.4 | 17155.9 | 40 | 10 | 3.83 | 31.63 | 83.05 | −0.325 |
| X76043.1 | 17125.9 | 39 | 11 | 3.90 | 31.77 | 82.94 | −0.345 |
| XM_002063468.1 | 17036.9 | 37 | 11 | 3.99 | 34.09 | 87.22 | −0.220 |
| AB180453.1 | 17262.2 | 42 | 13 | 4.05 | 37.38 | 84.12 | −0.322 |
| XM_001867954.1 | 18057.9 | 40 | 11 | 3.99 | 28.80 | 83.60 | −0.343 |
| AF432912.1 | 17304.2 | 42 | 13 | 4.01 | 38.57 | 84.84 | −0.457 |
| NM_001205104 | 17720.5 | 45 | 12 | 3.84 | 34.45 | 85.82 | −0.394 |
The alignment of the Per a 6 allergen and its
homologs is shown in Fig. 1. The
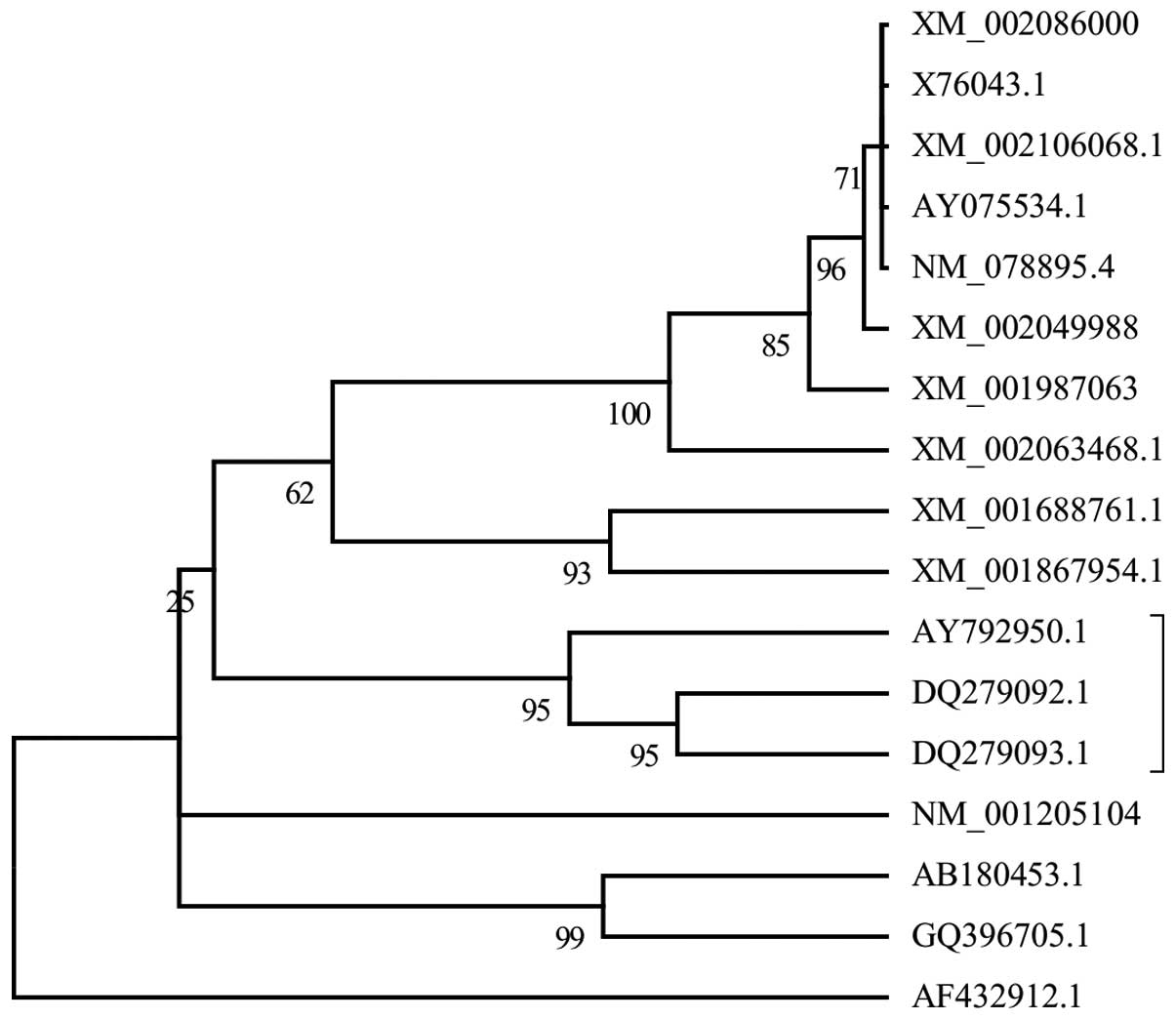
evolutionary tree (Fig. 2) was
inferred by the NJ method. Phylogenetic analysis showed that Bla g
6.01 (DQ279092.1) and Bla g 6.02 (DQ279093.1) were two related
sequences that were similar to the Per a 6 allergen. Bla g 6.01 and
Bla g 6.02 are two cockroach isoallergens identified from
Blatella germanica, with Bla g 6.01 exhibiting 93% amino
acid identity to Bla g 6.02. In the present study, the B- and
T-cell epitopes of the Per a 6 and Bla g 6.01 allergens were
investigated.
Epitope prediction
B-cell epitope prediction
Surface accessibility and fragment flexibility are
important features for predicting antigenic epitopes. In addition,
the existence of regions with high hydrophobicity provides strong
evidence for epitope identification. As shown in Figs. 3 and 4, the hydrophilic regions occupied most
of the sequence, indicating high hydrophilicity of the Per a 6 and
Bla g 6.01 allergens. Furthermore, the frequent appearance of the
surface and flexibility regions indicated that the Per a 6 and Bla
g 6.01 sequences were flexible, stretched and easily exposed to the
outside. The stretched structure was further validated by the
results of the flexibility and surface accessibility analysis. The
antigenic index directly showed the epitope-forming capacity of the
Per a 6 and Bla g 6.01 sequences. Most of the sequences exhibited a
high antigenic index, suggesting a high tendency of Per a 6 and Bla
g 6.01 to form epitopes. Based on these sequence properties, the
final regions predicted by DNAStar Protean were obtained as shown
in Tables II and III. Tables II and III also show the predicted results
obtained using the BPAP system and the BepiPred 1.0 server. The
final potential epitopes of the Per a 6 and Bla g 6.01 allergens
were selected on the basis of the results from these two tools and
those from the DNAStar Protean system. The ultimate results of the
three immunoinformatics tools predicted seven peptides (1–7, 19–26,
42–46, 81–86, 97–102, 117–122 and 129–140) to be the potential
linear B-cell epitopes of the Per a 6 allergen, and seven peptides,
1–7, 19–26, 55–63, 81–84, 97–102, 117–121 and 131–140, were
predicted as potential linear B-cell epitopes of the Bla g 6.01
allergen. The amino acid sequences are shown in Tables IV and V.
 | Table IILocation of the B-cell epitopes of
the Per a 6 allergen predicted by three immunoinformatics
tools. |
Table II
Location of the B-cell epitopes of
the Per a 6 allergen predicted by three immunoinformatics
tools.
| Tools | Location of the
prediction results |
|---|
| DNAStar
Protean | 1–7, 17–27, 42–48,
81–89, 96–102, 114–123, 129–140 |
| BPAP | 5–16, 32–41, 49–55,
64–78, 103–113 |
| BepiPred 1.0
server | 1–7, 19–26, 42–46,
81–86, 97–103, 117–122, 131–140 |
 | Table IIILocation of the B-cell epitopes of
the Bla g 6.01 allergen predicted by three immunoinformatics
tools. |
Table III
Location of the B-cell epitopes of
the Bla g 6.01 allergen predicted by three immunoinformatics
tools.
| Tools | Location of the
prediction results |
|---|
| DNAStar
Protean | 1–7, 17–27, 42–47,
55–65, 81–91, 95–102, 113–122, 129–140 |
| BPAP | 5–15, 23–29, 32–41,
48–54, 65–78, 107–113 |
| BepiPred 1.0
server | 1–7, 19–26, 55–63,
81–84, 97–103, 117–121, 131–140 |
 | Table IVB-cell epitopes of the Per a 6
allergen predicted by combined immunoinformatics methods. |
Table IV
B-cell epitopes of the Per a 6
allergen predicted by combined immunoinformatics methods.
| Number | Position | Length of sequence
(amino acids) | Sequence |
|---|
| Peptide 1 | 1–7 | 7 | MDELPDE |
| Peptide 2 | 19–26 | 8 | FDREKNGF |
| Peptide 3 | 42–46 | 5 | PLDDD |
| Peptide 4 | 81–86 | 6 | AEAMQQ |
| Peptide 5 | 97–102 | 6 | KEGNGY |
| Peptide 6 | 117–122 | 6 | DKLTNE |
| Peptide 7 | 129–140 | 12 | EEIDSDGSGTVD |
 | Table VB-cell epitopes of the Bla g 6
allergen predicted by combined immunoinformatics methods. |
Table V
B-cell epitopes of the Bla g 6
allergen predicted by combined immunoinformatics methods.
| Number | Position | Length of sequence
(amino acids) | Sequence |
|---|
| Peptide 1 | 1–7 | 7 | MDELPDE |
| Peptide 2 | 19–26 | 8 | FDREKKGC |
| Peptide 3 | 55–63 | 9 | VDADGSGEL |
| Peptide 4 | 81–84 | 4 | AEAM |
| Peptide 5 | 97–102 | 6 | KEGNGY |
| Peptide 6 | 117–121 | 5 | DKITA |
| Peptide 7 | 131–140 | 10 | IDSDGSGTVD |
T-cell epitope prediction
Net-MHCIIpan-2.0 and NetMHCII-2.2 were used to
identify significant core 9-mer peptides in the Per a 6 and Bla g
6.01 allergens. All estimations were based on the HLA-DRB1 alleles
found in the study cohort. For HLA-DQ alleles, only DQB1 0201,
0402, 0501 and 0602 were used for epitope prediction. In the
overall analysis using Net-MHCIIpan-2.0 and NetMHCII-2.2, Per a 6
was predicted to have nine strongly binding 9-mer core epitope
sequences (IC50 <50 nM): Peptides 7–15, 35–43, 65–73,
66–74, 68–76, 102–110, 121–129, 123–131, 142–150 (Table VI). The Per a 6 allergen was also
predicted to have 28 weakly binding 9-mer core epitope sequences
(50 nm <IC50 <500 nm) (Table VI). The Bla g 6.01 allergen was
predicted to have 34 significant nonamer core epitope sequences.
Nine of them were strong binders and comprised peptides 8–16,
35–43, 50–58, 68–76, 75–83, 84–92, 88–96, 89–97 and 142–150
(Table VII). The remaining
peptides were weak binders and are shown in Table VII.
 | Table VILocation of the T-cell epitopes of
the Per a 6 allergen predicted by Net-MHCIIpan-2.0 and
NetMHCII-2.2. |
Table VI
Location of the T-cell epitopes of
the Per a 6 allergen predicted by Net-MHCIIpan-2.0 and
NetMHCII-2.2.
| Per a 6
allergen |
|---|
| Strong-binding core
peptides | 7–15, 35–43, 65–73,
66–74, 68–76, 102–110, 121–129, 123–131, 142–150 |
| Weak-binding core
peptides | 9–17, 12–20, 26–34,
27–35, 28–36, 31–39, 32–40, 33–41, 36–44,47–55, 48–56, 51–59,
63–71, 64–72, 69–77, 75–83, 84–92, 86–94, 87–95, 88–96, 94–102,
95–103, 100–108, 103–111, 107–115, 111–119, 120–128, 140–148 |
 | Table VIILocation of the T-cell epitopes of
the Bla g 6 allergen predicted by Net-MHCIIpan-2.0 and
NetMHCII-2.2. |
Table VII
Location of the T-cell epitopes of
the Bla g 6 allergen predicted by Net-MHCIIpan-2.0 and
NetMHCII-2.2.
| Bla g 6.01
allergen |
|---|
| Strong-binding core
peptides | 8–16, 35–43, 50–58,
68–76, 75–83, 84–92, 88–96, 89–97, 142–150 |
| Weak-binding core
peptides | 4–12, 10–18, 12–20,
28–36, 24–32, 26–34, 41–49, 45–53, 48–56, 51–59, 65–73, 69–77,
70–78
73–81, 76–84, 77–85, 86–94, 95–103, 100–108, 102–110, 107–114,
108–116, 111–119, 119–127, 139–147 |
Discussion
The prevalence and morbidity of allergies, such as
asthma, rhinitis and eczema, in children have been increasing over
recent decades. This is currently a major concern for the
international medical community and cannot solely be attributed to
the improved diagnosis of these diseases (33). Conventional immunotherapy for
allergies using crude allergen extracts has been shown to have a
high risk of anaphylaxis. One alternative approach is the use of
peptide fragments corresponding to the B- and T-cell epitopes of
the allergen (34). In a previous
study, overlapping synthetic peptides were used to validate the
IgE-binding capacity. Although this method decreases the
possibility of missed epitopes, it requires the synthesis of large
numbers of peptides, which is time-consuming and costly (35). In silico prediction has
become a common and useful tool for selecting epitopes from
immunologically relevant proteins, and can save the time and cost
of peptide synthesis (36).
Per a 6 is a minor cockroach allergen from
Periplaneta americana, while Bla g 6 is an allergen from
Blatella germanica with a sensitization prevalence of 14%
among patients with cockroach allergy. Bla g 6 consists of three
isoallergens: Bla g 6.01, 6.02 and 6.03. Bla g 6.01 shows 93% amino
acid identity to Bla g 6.02 and 68% identity to Bla g 6.03. The
evolutionary tree in the present study (Fig. 2) showed that Bla g 6.01 and 6.02
are homologs of Per a 6 (11). To
the best of our knowledge, the present study is the first to
predict B- and T-cell epitopes from the Per a 6 and Bla g 6.01
allergens. A previous study showed that the prediction of B-cell
epitopes using the bioinformatics approach correlated well with the
experimental approach (37). The
prediction of B-cell epitopes among protein sequences has been
enabled by the development of numerous algorithms that are based on
certain amino acid properties, including hydrophilicity,
antigenicity, segmental mobility, flexibility and accessibility
(38). In the present study, the
linear B-cell epitopes of the Per a 6 and Bla g 6.01 allergens were
predicted using three sequence-based tools (the DNAStar Protean
system, BPAP and the BepiPred 1.0 server). Seven B-cell epitopes of
Per a 6 and seven epitopes of Bla g 6 were investigated. The
alignment results (Fig. 1) showed
that all the peptides of Per a 6 and Bla g 6 were located in
similar positions, with exception of peptide three. This indicated
that Per a 6 shares a cross-reactive epitope with Bla g 6. Peptide
three of Per a 6 was located at position 42–46, while that of Bla g
6 was located at 55–63, indicating that peptide three may represent
a distinction between the epitopes of these two cockroach
allergens.
In recent years, certain algorithms have
substantially improved in their accuracy to predict T-cell
epitopes. However, most algorithms have targeted HLA-DR molecules,
not HLA-DP or HLA-DQ molecules, despite the importance of HLA-DP
and HLA-DQ molecules for antigen presentation. NetMHCpan-2.0 was
recently shown to have a per-allele mean accuracy of 0.854 (1.0
being 100% accurate and 0.5 of no significance) (39). In another study, the per-allele
mean accuracy of the NN-align method was 0.882 (31). In the present study,
Net-MHCIIpan-2.0 and NetMHCII-2.2 were used to predict the core
9-mer T-cell epitopes in the Per a 6 and Bla g 6 allergens.
Combining the results obtained with Net-MHCIIpan-2.0 and
NetMHCII-2.2, a total of 37 and 34 epitopes were predicted for the
Per a 6 and Bla g 6 allergens, respectively. Nine epitopes from the
Per a 6 allergen and nine from Bla g 6 allergen were strongly
binding, with an IC50<50 nm.
In conclusion, the present study predicted B- and
T-cell epitopes of the Per a 6 allergen and its homolog, the Bla g
6.01 allergen in Blatella germanica, using an in
silico method. The results may be used to benefit allergen
immunotherapies and reduce the frequency of allergic reactions.
However, the accuracy of these epitopes requires confirmation in
further experiments.
Acknowledgements
The present study was sponsored by the National
Natural Science Foundation of China (nos. 31340073, 81273274,
81373128, 810001329 and 30972822), the Great Project (no.
2011ZX08011-005) from the Major Program of National Science and
Technology of China and the Special Research Project (no.
201300000159) from the Science and Information Technology of
Guangzhou. National Major Scientific and Technological Special
Project for ‘Significant New Drugs Development’
(2011ZX09302-003-02), and a project funded by the Priority Academic
Development Program of Jiangsu Higher Education Institutions.
References
|
1
|
Bernton HS and Brown H: Insect allergy -
preliminary studies of the cockroach. J Allergy. 35:506–513. 1964.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Arruda LK, Vailes LD, Ferriani VP, Santos
AB, Pomés A and Chapman MD: Cockroach allergens and asthma. J
Allergy Clin Immunol. 107:419–428. 2001. View Article : Google Scholar
|
|
3
|
Sun BQ, Lai XX, Gjesing B, Spangfort MD
and Zhong NS: Prevalence of sensitivity to cockroach allergens and
IgE cross-reactivity between cockroach and house dust mite
allergens in Chinese patients with allergic rhinitis and asthma.
Chin Med J (Engl). 123:3540–3544. 2010.PubMed/NCBI
|
|
4
|
He S, Zhang Z, Zhang H, et al: Analysis of
properties and proinflammatory functions of cockroach allergens Per
a 1.01s. Scand J Immunol. 74:288–295. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Pan QR, Wang SM, Shang HS and Chew FT:
Identification and characterization of Per a 2, the Bla g 2
allergen homologue from American cockroach (Periplaneta
americana). J Allergy Clin Immunol. 117:S1152006. View Article : Google Scholar
|
|
6
|
Tan YW, Chan SL, Ong TC, et al: Structures
of two major allergens, Bla g 4 and Per a 4, from cockroaches and
their IgE binding epitopes. J Biol Chem. 284:3148–3157. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Khantisitthiporn O, Sookrung N,
Tungtrongchitr A, et al: Native troponin-T of the American
cockroach (CR), Periplaneta americana, binds to IgE in sera
of CR allergic Thais. Asian Pac J Allergy Immunol. 25:189–197.
2007.
|
|
8
|
Asturias JA, Gómez-Bayón N, Arilla MC, et
al: Molecular characterization of American cockroach tropomyosin
(Periplaneta americana allergen 7), a cross-reactive
allergen. J Immunol. 162:4342–4348. 1999.PubMed/NCBI
|
|
9
|
Sookrung N, Chaicumpa W, Tungtrongchitr A,
et al: Periplaneta americana arginine kinase as a major
cockroach allergen among Thai patients with major cockroach
allergies. Environ Health Perspect. 114:875–880. 2006. View Article : Google Scholar
|
|
10
|
Sudha VT, Arora N, Gaur SN, Pasha S and
Singh BP: Identification of a serine protease as a major allergen
(Per a 10) of Periplaneta americana. Allergy. 63:768–776.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sookrung N and Chaicumpa W: A revisit to
cockroach allergens. Asian Pac J Allergy Immunol. 28:95–106.
2010.PubMed/NCBI
|
|
12
|
Kang BC, Johnson J, Morgan C and Chang JL:
The role of immunotherapy in cockroach asthma. J Asthma.
25:205–218. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Srivastava D, Gaur SN, Arora N and Singh
BP: Clinico-immunological changes post-immunotherapy with
Periplaneta americana. Eur J Clin Invest. 41:879–888. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sharma V, Singh BP, Gaur SN, Pasha S and
Arora N: Bioinformatics and immunologic investigation on B and T
cell epitopes of Cur I 3, a major allergen of Curvularia
lunata. J Proteome Res. 8:2650–2655. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang HW, Lin YC, Pai TW and Chang HT:
Prediction of B-cell linear epitopes with a combination of support
vector machine classification and amino acid propensity
identification. J Biomed Biotechnol. 2011:4328302011.
|
|
16
|
Jiminez-Lopez JC, Kotchoni SO,
Rodríguez-García MI and Alché JD: Structure and functional features
of olive pollen pectin methylesterase using homology modeling and
molecular docking methods. J Mol Model. 18:4965–4984. 2012.
View Article : Google Scholar
|
|
17
|
Nielsen M, Lund O, Buus S and Lundegaard
C: MHC class II epitope predictive algorithms. Immunology.
130:319–328. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Larkin MA, Blackshields G, Brown NP, et
al: Clustal W and Clustal X version 2.0. Bioinformatics.
23:2947–2948. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kumar S, Nei M, Dudley J and Tamura K:
MEGA: a biologist-centric software for evolutionary analysis of DNA
and protein sequences. Brief Bioinform. 9:299–306. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hopp TP and Woods KR: Prediction of
protein antigenic determinants from amino acid sequences. Proc Natl
Acad Sci USA. 78:3824–3828. 1981. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kyte J and Doolittle RF: A simple method
for displaying the hydropathic character of a protein. J Mol Biol.
157:105–132. 1982. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Karplus PA and Schulz GE: Prediction of
chain flexibility in proteins. Naturwissenschaften. 72:212–213.
1985. View Article : Google Scholar
|
|
23
|
Emini EA, Hughes JV, Perlow D and Boger J:
Induction of hepatitis A virus-neutralizing antibody by a
virus-specific synthetic peptide. J Virol. 55:836–839.
1985.PubMed/NCBI
|
|
24
|
Jameson B and Wolf H: The antigenic index:
a novel algorithm for predicting antigenic determinants. Comput
Appl Biosci. 4:181–186. 1988.PubMed/NCBI
|
|
25
|
Kolaskar A and Tongaonkar PC: A
semi-empirical method for prediction of antigenic determinants on
protein antigens. FEBS Lett. 276:172–174. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Larsen JE, Lund O and Nielsen M: Improved
method for predicting linear B-cell epitopes. Immunome Res.
2:22006. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zheng LN, Lin H, Pawar R, Li ZX and Li MH:
Mapping IgE binding epitopes of major shrimp (Penaeus
monodon) allergen with immunoinformatics tools. Food Chem
Toxicol. 49:2954–2960. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yang X and Yu X: An introduction to
epitope prediction methods and software. Rev Med Virol. 19:77–96.
2009. View
Article : Google Scholar : PubMed/NCBI
|
|
29
|
Rammensee HG, Friede T and Stevanoviíc S:
MHC ligands and peptide motifs: first listing. Immunogenetics.
41:178–228. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Nielsen M and Lund O: NN-align. An
artificial neural network-based alignment algorithm for MHC class
II peptide binding prediction. BMC Bioinformatics. 10:2962009.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wang P, Sidney J, Kim Y, et al: Peptide
binding predictions for HLA DR, DP and DQ molecules. BMC
Bioinformatics. 11:5682010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Pascal M, Konstantinou GN, Masilamani M,
Lieberman J and Sampson HA: In silico prediction of Ara h 2 T cell
epitopes in peanut-allergic children. Clin Exp Allergy. 43:116–127.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Baena-Cagnani CE, Serra H, Teijeiro A and
Croce JS: Prevention of allergy and asthma. Clin Exp Allergy Rev.
3:51–57. 2003. View Article : Google Scholar
|
|
34
|
Moldaver D and Larché M: Immunotherapy
with peptides. Allergy. 66:784–791. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Lin J, Bardina L, Shreffler WG, et al:
Development of a novel peptide microarray for large-scale epitope
mapping of food allergens. J Allergy Clin Immunol. 124:315–322.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Li GF, Wang Y, Zhang ZS, et al:
Identification of immunodominant Th1-type T cell epitopes from
Schistosoma japonicum 28 kDa glutathione-S-transferase, a
vaccine candidate. Acta Biochim Biophys Sin (Shanghai). 37:751–758.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Nair S, Kukreja N, Singh BP and Arora N:
Identification of B cell epitopes of alcohol dehydrogenase allergen
of Curvularia lunata. PLoS One. 6:e200202011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Pomés A: Relevant B cell epitopes in
allergic disease. Int Arch Allergy Immunol. 152:1–11. 2010.
|
|
39
|
Nielsen M, Justesen S, Lund O, Lundegaard
C and Buus S: NetMHCIIpan-2.0 - Improved pan-specific HLA-DR
predictions using a novel concurrent alignment and weight
optimization training procedure. Immunome Res. 6:92010. View Article : Google Scholar : PubMed/NCBI
|