Introduction
Lung cancer is currently one of the most common
types of malignant cancers (1).
The incidence of lung cancer is evidently increasing in China
(2). Approximately 80% of all lung
cancer is non-small cell lung cancer (NSCLC) (3,4).
Numerous studies have focused on improving diagnosis and therapy,
but 30–40% patients with NSCLC still have a poor prognosis. The
most common feature of malignancy is invasion, which is responsible
for the low 5-year survival rates. Therefore, determining the
mechanism underlying the association between miR-30c and invasion
would facilitate the understanding of the progression of NSCLC and
thus contribute to developing novel therapeutic agents.
MicroRNAs (miRNAs) had been identified as having
important roles in controlling the expression of downstream target
genes in various biological processes (5–7). A
number of studies have suggested that miRNAs may regulate the
proliferation, apoptosis, cell cycle and invasion of cancer cells
(8,9). The miR-30 family (miR-30a/b/c/d/e/f)
has also been reported in various diseases, including breast cancer
(10), retinal pigment epithelial
cell cancer (11), glioma
(12) and osteoblastic cancer
(13).
Epithelial-to-mesenchymal transition (EMT) has a
pivotal role in the invasion of various cancer types by the
transformation of polarized and adherent epithelial cells into
motile and invasive mesenchymal cells (14,15).
Numerous transcription factors involved in EMT, including Snail and
Twist, upregulate the expression of mesenchymal markers, such as
vimentin, collagen and fibronectin and downregulate the expression
of epithelial markers, including E-cadherin. A breakdown of tight
junctions is involved in the loss of epithelial markers and
acquisition of mesenchymal makers (16–18).
The present study aimed to examine the underlying
mechanism of the association between miR-30c and invasion in NSCLC,
in order to provide further evidence to facilitate improvement of
the therapeutic strategies for this disease.
Materials and methods
Clinical samples
A total of 85 patients with NSCLC that had undergone
routine surgery at The First Affiliated Hospital of Nanjing Medical
University (Nanjing, China) between May 2010 and November 2012 were
selected to participate in this study. The NSCLC samples and the
adjacent lung tissues obtained from the 85 patients were collected,
immediately snap frozen in liquid nitrogen and stored at −80°C
until RNA extraction. The tumors were classified according to World
Health Organization classification (19). The present study was approved by
the Ethical Committee of The First Affiliated Hospital of Nanjing
Medical University and every patient provided written informed
consent.
Cell culture
The A549 cell line (American Type Culture
Collection, Manassas, VA, USA) was employed for the present study
and was cultured in RPMI-1640 medium with 10% fetal bovine serum
(Invitrogen, Carlsbad, CA, USA) and penicillin (100 U/ml). The
cells were cultured at 37°C with 5% CO2.
Isolation of total RNA and quantitative
polymerase chain reaction (qPCR)
Total RNA was extracted from collected tissues using
TRIzol reagent (Invitrogen) and then mRNA was reverse transcribed
to cDNA. The stem-loop primer for miR-30c was
5′-GTCGTATCCAGTGCAGGGTCCGAGTATTCGCACTGGATACGACGCTGA-3′. U6 small
nuclear RNA was used for normalization. The PCR reactions were
performed with the following primers: Forward:
5′-GCCGCTGTAAACATCCTACACT-3′ and reverse: 5′-GTGCAGGGTCCGAGGT-3′
for hsa-miR-30c; and forward: 5′-CTCGCTTCGGCAGCACA-3′ and reverse:
5′-AACGCTTCACGAATTTGCGT-3′ for U6. Reaction conditions were as
follows: 37°C for 15 min and 85°C for 5 sec. Unused reaction
products were stored at 4°C. qPCR was performed using the ABI 7500
Fast Real-Time PCR system (Applied Biosystems, Carlsbad, CA,
USA).
Wound healing assay
The cells were plated onto 6-well plates and
cultured with RPMI-1640 medium. Following 24 h, the cells were
wounded with a pipette tip. Serum-free RPMI-1640 medium was added
and wound closure was observed for 24 h using an XSP-4C microscope
(Shanghai Changfang Optical Instrument Co. Ltd., Shanghai,
China).
Transwell assay
The cell motility was measured using an 8-μm-pore
polycarbonate membrane Boyden chamber insert in a Transwell
apparatus (Millipore, Billerica, MA, USA). The transfected cells
were treated with trypsin/EDTA solution and washed once with
serum-containing RPMI-1640 medium. A total of 1×105
cells in 0.2 ml serum-free RPMI-1640 medium were seeded onto a
Transwell apparatus. RPMI-1640 containing 10% fetal bovine serum
(600 μl) was added to the lower chamber. An invasion assay was
conducted following the same procedure, with the exception that the
filters of the Transwell chambers were coated with 45 μg Matrigel
(BD Biosciences, San Jose, CA, USA). Following incubation of the
cells for 24 h at 37°C in a 5% CO2 incubator, the cells
on the top surface of the insert were removed by wiping with a
cotton swab. The cells that invaded to the bottom surface of the
insert were fixed in the 100% precooling methanol for 10 min,
stained in 0.5% crystal violet for 30 min, then rinsed in
phosphate-buffered saline (PBS) and subjected to microscopic
inspection. The values for invasion were obtained by counting three
fields per membrane and represented the average of three
independent experiments.
Western blot analysis
The total proteins were prepared from the
established cells, quantities using a protein assay (bicinchoninic
acid method; Beyotime, Shanghai, China). The proteins were
fractionated by sodium dodecyl sulfate-polyacrylamide gel
electrophoresis (SDS-PAGE), transferred to polyvinylidene fluoride
(PVDF) membrane (Millipore), blocked in 5% dry milk at room
temperature for 1 h and immunostained with antibodies at 4°C
overnight using anti-E-cadherin, anti-Snail, anti-vimentin
(1:1,000; Dizhao, Nanjing, China) and anti-GAPDH (1:5,000; Kangchen
KangChen Bio-Tech, Shanghai, China). All of the results were
visualized through a chemiluminescent detection system (Pierce ECL
Substrate western blot detection system; Thermo Scientific,
Pittsburgh, PA, USA) and then exposed in Molecular Imager ChemiDoc
XRS System (Bio-Rad, Hercules, CA, USA). The integrated density of
the band was quantified by ImageJ software (Bio-Rad).
Transfection
The A549 cells were plated in 6-well plates
(6×105 cells/well) and 100 nm of the miR-30c mimic or
100 nm miRNA mimic control were transfected into the A549 cells,
while 100 nm of the miR-30c inhibitor (anti-miR-30c) or 100 nm
miRNA inhibitor control were transfected into the A549 cells, using
Lipofectamine 2000 (Invitrogen Life Technologies, Grand Island, NY,
USA) according to the manufacturer’s instructions. The miR-30c
mimic, miRNA mimic control, miR-30c inhibitor and miRNA inhibitor
control were purchased from Shanghai GenePharma Co., Ltd.
(Shanghai, China).
Statistical analysis
The 2−ΔΔCt method was used to analyze the
results of qPCR in all of the experiments performed in the present
study. Statistical analysis was performed using STATA 11, and
presented with Graph Pad prism software (GraphPad Software, Inc.,
La Jolla, CA, USA). The results obtained from experiment in
vitro assays are presented as the mean ± standard error of the
mean from five separate experiments in triplicates per experiment,
and the data was analyzed by the Wilcoxon rank-sum (Mann-Whitney)
test. P<0.05 was considered to indicate a statistically
significant difference.
Results
miR-30c is reduced in human lung cancer
tissues
The expression of miR-30c was analyzed in lung
cancer samples (n=85) and adjacent lung tissues by qPCR. The
miR-30c expression was significantly lower in lung cancer tissues
than paraneoplastic tissues (P=0.007; Fig. 1). There was no positive correlation
with gender, age, smoking status, histological type or tumor size,
however, there was an evident correlation with tumor stage
(P=0.026) and metastasis (P=0.009; Table I). The aberrant expression level of
miR-30c suggested that miR-30c may have an important role in lung
cancer progression and development. Therefore, based on this
expression pattern, the A549 cell line was selected to verify the
effect of miR-30c.
 | Table IExpression level of miR-30c in lung
cancer and corresponding adjacent tissues. |
Table I
Expression level of miR-30c in lung
cancer and corresponding adjacent tissues.
| Factor | No. of samples | miR-30c
low-expression (<median) | miR-30c
high-expression (≥median) | P-value |
|---|
| Gender | | | | 0.950 |
| Male | 52 | 28 | 24 | |
| Female | 33 | 18 | 15 | |
| Age (years) | | | | 0.629 |
| <60 | 39 | 20 | 19 | |
| ≥60 | 46 | 26 | 20 | |
| Smoker | | | | 0.832 |
| No | 36 | 19 | 17 | |
| Yes | 49 | 27 | 22 | |
| Histological
type | | | | 0.805 |
| SC | 47 | 26 | 21 | |
| AC | 38 | 20 | 18 | |
| Tumor stage | | | | 0.026a |
| I–II | 39 | 16 | 23 | |
| III–IV | 46 | 30 | 16 | |
| Tumor size | | | | 0.047a |
| T1/T2 | 49 | 22 | 27 | |
| T3/T4 | 36 | 24 | 12 | |
| Metastasis | | | | 0.009b |
| No | 31 | 11 | 20 | |
| Yes | 54 | 35 | 19 | |
| Total | 85 | 46 | 39 | |
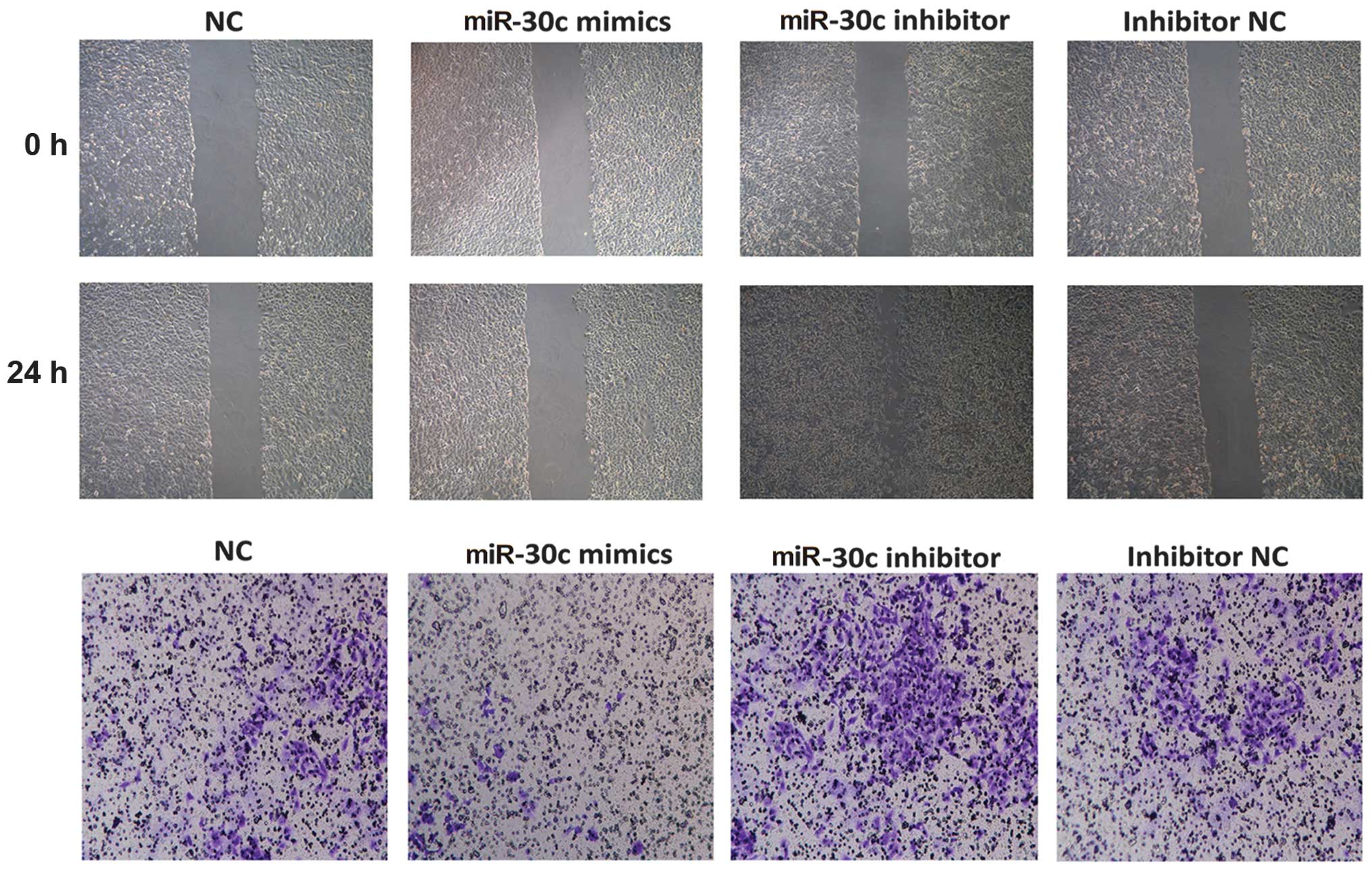
miR-30c regulated the invasion of A549
cells in vitro
To examine the mechanism underlying the effect of
miR-30c on the invasion in lung cancer, the A549 cells were
transfected with miR-30c mimics, NC mimics and miR-30c inhibitor
(anti-miR-30c) and inhibitor NC respectively. The transfection
efficiency was validated by qPCR (Fig.
1). The wound healing assay demonstrated that the
overexpression of miR-30c was able to suppress A549 cell healing,
while suppression of miR-30c increased cell healing (Fig. 2). Furthermore, the Matrigel
invasion assay demonstrated that overexpression of miR-30c
attenuated A549 cell invasion, whereas the suppression of miR-30c
reversed its effect (Fig. 2). The
results suggested that miR-30c inhibited invasion of the A549 cell
line in vitro.
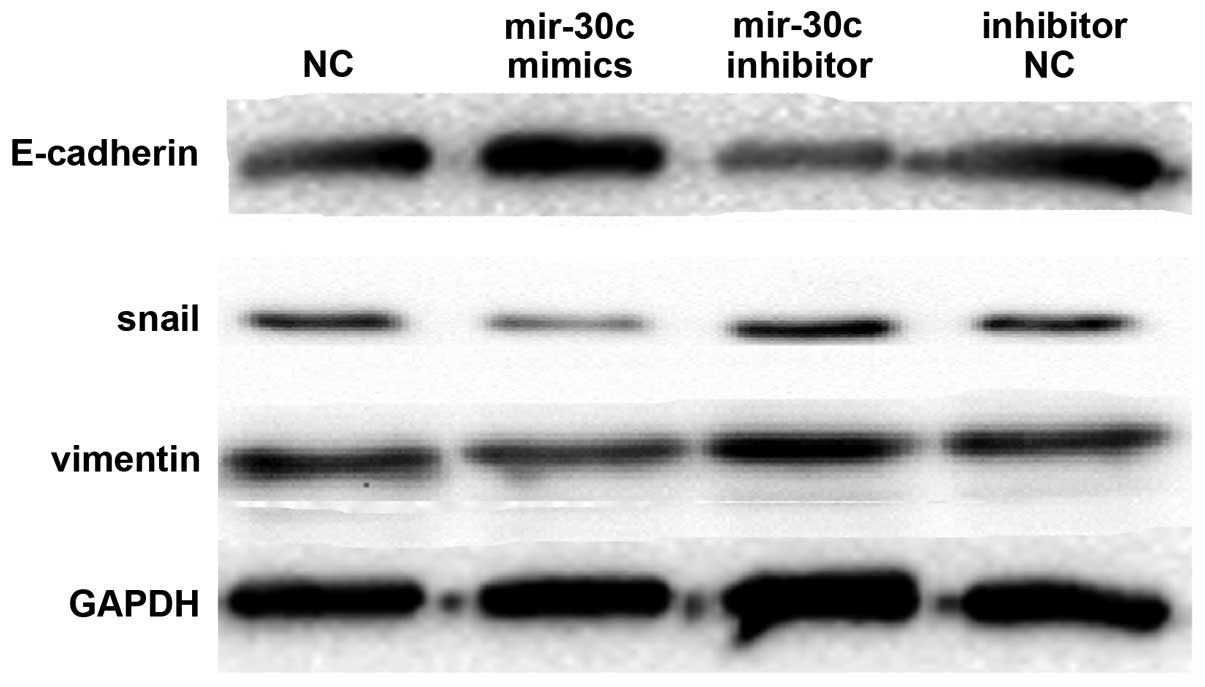
Down regulated expression of miR-30c
induces EMT
The A549 cells were transfected with miR-30c mimics,
NC mimics and miR-30c inhibitor (anti-miR-30c) and inhibitor NC to
examine whether miR-30c was involved in EMT. The epithelial marker
(E-cadherin) and mesenchymal markers (vimentin and Snail) were
investigated by western blot analysis. At a protein level,
upregulated miR-30c expression by miR-30c mimics resulted in
elevated E-cadherin expression and decreased vimentin and Snail
expression. In addition, suppression of miR-30c expression by the
miR-30c inhibitor resulted in decreased E-cadherin expression and
increased vimentin and Snail expression (Fig. 3). Therefore, it was concluded that
miR-30c contributed to regulating EMT marker expression in lung
cancer cell lines.
Discussion
The present results indicated that the expression of
miR-30c was decreased in lung cancer tissues (n=85), as compared
with the corresponding adjacent tissues. Aberrant expression of
miR-30c controlled the invasion of lung cancer cell lines in
vitro. Furthermore, it was also identified that the
overexpression of miR-30c led to elevated E-cadherin expression and
decreased vimentin and Snail expression. The downregulation of
miR-30c had the reverse effect. E-cadherin is an epithelial marker,
while vimentin and Snail are mesenchymal markers. These results
suggested that downregulation of miR-30c may promote lung cancer
invasion by inducing EMT.
Decreased E-cadherin and elevated vimentin and Snail
expression is a hallmark of EMT, which is a key process in cancer
invasion (19). Previously, EMT
has been identified to be associated with tumor invasiveness,
metastasis and prognosis (20,21).
Numerous studies established functional associations between
non-coding microRNAs and key effectors of EMT occurring in the
context of carcinogenesis and embryonic development, including
microRNA-200 (22,23), microRNA-10b (24) and microRNA-21 (25,26).
In addition to cancer progression, EMT contributes to chronic
epithelial injury (27), leading
to tissue fibrosis and organ failure (28,29).
In conclusion, compared with the adjacent tissues,
the mRNA expression level of miR-30c was decreased in lung cancer.
It was demonstrated that low expression of miR-30c promoted
invasion via inducing EMT in lung cancer. Furthermore, the
miR-30c-EMT pathway that was investigated may be exploited in a
therapeutic approach for the treatment of lung cancer in the
future.
Acknowledgements
The authors would like to thank Dr Junwei Tang for
help with reviewing the language of the manuscript.
References
|
1
|
Jemal A, Siegel R, Xu J and Ward E: Cancer
statistics, 2010. CA Cancer J Clin. 60:277–300. 2010. View Article : Google Scholar
|
|
2
|
Yang L, Parkin DM, Ferlay J, Li L and Chen
Y: Estimates of cancer incidence in China for 2000 and projections
for 2005. Cancer Epidemiol Biomarkers Prev. 14:243–250.
2005.PubMed/NCBI
|
|
3
|
Herbst RS, Heymach JV and Lippman SM: Lung
cancer. New Engl J Med. 359:1367–1380. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Brundage MD, Davies D and Mackillop WJ:
Prognostic factors in non-small cell lung cancer: a decade of
progress. Chest. 122:1037–1057. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhang J, Kong X, Li J, et al: MiR-96
promotes tumor proliferation and invasion by targeting RECK in
breast cancer. Oncol Rep. 31:1357–1363. 2013.PubMed/NCBI
|
|
6
|
Yang J, Zhao H, Xin Y and Fan L:
MicroRNA-198 inhibits proliferation and induces apoptosis of lung
cancer cells via targeting FGFR1. J Cell Biochem. 115:987–995.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang F, Xia J, Wang N and Zong H: miR-145
inhibits proliferation and invasion of esophageal squamous cell
carcinoma in part by targeting c-Myc. Onkologie. 36:754–758.
2013.PubMed/NCBI
|
|
8
|
Mita M: Regional differences of the clear
cells in the mouse epididymal duct: a histological study. Hokkaido
Igaku Zasshi. 61:909–920. 1986.(In Japanese).
|
|
9
|
Li H, Xu H, Shen H and Li H: microRNA-106a
modulates cisplatin sensitivity by targeting PDCD4 in human ovarian
cancer cells. Oncol Lett. 7:183–188. 2014.PubMed/NCBI
|
|
10
|
Ouzounova M, Vuong T, Ancey PB, et al:
MicroRNA miR-30 family regulates non-attachment growth of breast
cancer cells. BMC Genomics. 14:1392013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Haque R, Chun E, Howell JC, et al:
MicroRNA-30b-mediated regulation of catalase expression in human
ARPE-19 cells. PloS One. 7:e425422012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Quintavalle C, Donnarumma E, Iaboni M, et
al: Effect of miR-21 and miR-30b/c on TRAIL-induced apoptosis in
glioma cells. Oncogene. 32:4001–4008. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wu T, Zhou H, Hong Y, et al: miR-30 family
members negatively regulate osteoblast differentiation. J Biol
Chem. 287:7503–7511. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang Y, Wen M, Kwon Y, et al: CUL4A
induces epithelial-mesenchymal transition and promotes cancer
metastasis by regulating ZEB1 expression. Cancer Res. 74:520–531.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liu J, Ruan B, You N, et al:
Downregulation of miR-200a induces EMT phenotypes and CSC-like
signatures through targeting the β-catenin pathway in hepatic oval
cells. PloS One. 8:e794092013.PubMed/NCBI
|
|
16
|
Dong H, Xie L, Tang C, et al: Snail1
correlates with patient outcomes in E-cadherin-preserved
gastroesophageal junction adenocarcinoma. Clin Transl Oncol. Dec
20–2013.(Epub ahead of print).
|
|
17
|
Liu Y, Li H, Feng J, et al: Lin28 induces
epithelial-to-mesenchymal transition and stemness via
downregulation of let-7a in breast cancer cells. PloS One.
8:e830832013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Bao YX, Cao Q, Yang Y, et al: Expression
and prognostic significance of golgiglycoprotein73 (GP73) with
epithelial-mesenchymal transition (EMT) related molecules in
hepatocellular carcinoma (HCC). Diagn Pathol. 8:1972013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kitamura K, Seike M, Okano T, et al:
MiR-134/487b/655 cluster regulates TGF-β-induced
epithelial-mesenchymal transition and drug resistance to gefitinib
by targeting MAGI2 in lung adenocarcinoma cells. Mol Cancer Ther.
13:444–453. 2014.PubMed/NCBI
|
|
20
|
Guo S, Xu X, Tang Y, et al: miR-15a
inhibits cell proliferation and epithelial to mesenchymal
transition in pancreatic ductal adenocarcinoma by down-regulating
Bmi-1 expression. Cancer Lett. 344:40–46. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yamada S, Fuchs BC, Fujii T, et al:
Epithelial-to-mesenchymal transition predicts prognosis of
pancreatic cancer. Surgery. 154:946–954. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Paterson EL, Kazenwadel J, Bert AG, et al:
Down-regulation of the miRNA-200 family at the invasive front of
colorectal cancers with degraded basement membrane indicates EMT is
involved in cancer progression. Neoplasia. 15:180–191. 2013.
|
|
23
|
Bai JX, Yan B, Zhao ZN, et al: Tamoxifen
represses miR-200 microRNAs and promotes epithelial-to-mesenchymal
transition by up-regulating c-Myc in endometrial carcinoma cell
lines. Endocrinology. 154:635–645. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ouyang H, Gore J, Deitz S and Korc M:
MicroRNA-10b enhances pancreatic cancer cell invasion by
suppressing TIP30 expression and promoting EGF and TGF-β actions.
Oncogene. Oct 7–2013.(Epub ahead of print).
|
|
25
|
Brønnum H, Andersen DC, Schneider M, et
al: miR-21 promotes fibrogenic epithelial-to-mesenchymal transition
of epicardial mesothelial cells involving Programmed Cell Death 4
and Sprouty-1. PloS One. 8:e562802013.
|
|
26
|
Han M, Wang Y, Liu M, et al: MiR-21
regulates epithelial-mesenchymal transition phenotype and
hypoxia-inducible factor-1alpha expression in third-sphere forming
breast cancer stem cell-like cells. Cancer Sci. 103:1058–1064.
2012. View Article : Google Scholar
|
|
27
|
Vitalone MJ, Naesens M, Sigdel T, et al:
The dual role of epithelial-to-mesenchymal transition in chronic
allograft injury in pediatric renal transplantation.
Transplantation. 92:787–795. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
López-Novoa JM and Nieto MA: Inflammation
and EMT: an alliance towards organ fibrosis and cancer progression.
EMBO Mol Med. 1:303–314. 2009.PubMed/NCBI
|
|
29
|
Mucsi I and Rosivall L:
Epithelial-mesenchymal transition in renal tubular cells in the
pathogenesis of progressive tubulo-interstitial fibrosis. Acta
Physiol Hung. 94:117–131. 2007. View Article : Google Scholar : PubMed/NCBI
|