Introduction
Congenital heart defects (CHD), which include
malformations of the heart or great vessels, are the most common
group of major birth defects, with an incidence of 5–8 per 1,000
live births (1). MicroRNAs
(miRNAs) that contribute to cardiac development have been
identified and can be used as novel biomarkers and therapeutic
targets for CHD (2), as previously
demonstrated with non-small cell lung cancer (3). MicroRNA-19b (miR-19b) is part of the
miR-17–92 cluster, which encodes miR-17, miR-18a, miR-19a, miR-19b,
miR-20a and miR-92a-1. The miR-17–92 cluster is required to induce
cardiomyocyte proliferation in postnatal and adult hearts (4). A number of studies have shown that
the miR-17–92 cluster contributes to the development of the heart,
lungs, blood vessels and immune system (5). A previous study has observed specific
changes in miRNA abundance and activity in a broad range of human
aging models and suggested the use of miR-17, miR-19b, miR-20a and
miR-106 as novel biomarkers of cellular aging (6).
P19 cells, isolated from an experimental
embryo-derived mouse teratocarcinoma, differentiate into embryonic
myocardial cells when exposed to dimethylsulfoxide (DMSO) (7). Therefore, they can be used to
investigate cardiac-specific transcription factors and upstream
signaling pathways during cardiac cell differentiation (8–10).
In addition, P19 cells are an excellent model system for studying
the regulation of myocardial electrophysiological differentiation
at the molecular and functional levels (11).
The Wnt signaling pathway performs a number of
functions during cardiogenesis (12). Early activation of Wnt/β-catenin
signaling promotes cardiac differentiation in zebrafish embryos and
mouse embryonic stem cells. Activation of Wnt/β-catenin at later
stages results in the repression of cardiac differentiation
(13). However, whether miR-19b
knockdown affects the cardiac lineage commitment and
differentiation through Wnt/β-catenin signaling remains to be
determined.
As P19 cells can differentiate into cardiomyocytes,
the present study investigated the underlying mechanisms of heart
development by analyzing the proliferation, apoptosis and
differentiation of P19 miR-19b-knockdown cells.
Materials and methods
Cell culture and induction of
differentiation
P19 cells were obtained from the American Type
Culture Collection (ATCC, Manassas, VA, USA). The cells were
cultured in modified Eagle’s medium (α-MEM; Gibco-BRL, Grand
Island, NY, USA) containing 10% fetal bovine serum (FBS;
Gibco-BRL), 100 mg/ml streptomycin and 100 U/ml penicillin in a 5%
CO2 atmosphere at 37°C. To induce cardiac
differentiation, the cells were cultivated in 10 ml α-MEM
supplemented with 10% FBS, 100 U/ml penicillin, 100 mg/ml
streptomycin and 1% DMSO (Sigma, St. Louis, MO, USA) in 10-cm
bacterial dishes in a 5% CO2 atmosphere at 37°C from
days 0 to 4. On day 4, the embryoid bodies were transferred to 6-cm
cell culture flasks with complete medium and cultured for an
additional 8 days. Cells were harvested on differentiation days 0,
4, 8, 10 and 12. Morphological changes in the P19 cells were
examined under an inverted microscope (Nikon Eclipse TE300; Nikon,
Tokyo, Japan) equipped with phase-contrast objectives and a digital
camera (E4500; Nikon). To investigate the differentiation process
in P19 cells, quantitative polymerase chain reaction (qPCR) was
used to identify the expression levels of cardiac troponin T
(cTnT), GATA4 and NKX2.5 during differentiation.
MiRNA transfection and establishment of
stable cell lines
Lipofectamine 2000 was used to transfect the
plasmids (pGLV3/H1/eGFP/Puro-miR-19b-3p-inhibitor sponge and
pGLV3/H1/eGFP/Puro-miR-vector; GenePharma, Shanghai, China) into
P19 cells. Puromycin (Invitrogen, Carlsbad, CA, USA), which kills
untransduced cells upon addition of the minimum concentration, was
used to select stably transduced cells.
CCK-8 assay
Cell Counting kit-8 (CCK-8; Dojindo, Kumamoto,
Japan) was used to assess cell growth according to the
manufacturer’s instructions. The stable cell lines which were
established with the miR-19b silencing expression plasmid or vector
were seeded in 96-well plates and maintained in α-MEM supplemented
with 10% FBS, 100 U/ml penicillin and 100 mg/ml streptomycin for
seven consecutive days. In brief, the CCK-8 solution (10% of the
medium, 10 μl) was added to each well and incubated for 1 h prior
to analysis with a microplate reader (DNM-9602, Beijing Perlong
Medical Instrument Ltd, Beijing, China) with the absorbance
measured at a wavelength of 450 nm. The results were plotted as the
mean ± standard deviation of three separate experiments, with three
determinations per experiment for each experimental condition.
Cell cycle assay
MiR-19b silenced or vector control stable P19 cells
were plated in α-MEM with 10% FBS, 100 U/ml penicillin and 100
mg/ml streptomycin. The cells were serum deprived for 24 h to
synchronize them and, following replacement of the starvation
medium with complete medium, were harvested using trypsin/EDTA,
washed twice with phosphate-buffered saline (PBS), fixed in 70%
ethanol at −20°C overnight and then stained with 500 ml propidium
iodide (PI) solution (100 mg/ml RNase and 50 mg/ml PI in 1X PBS).
Cell cycle analysis was initiated at multiple time points (0, 8,
16, 24 and 32 h). BD FACScan and Cell Quest software (BD
Biosciences, San Jose, CA, USA) were used to analyze the labeled
cultured cells.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted from P19 cells using the
TRIzol reagent and cDNA was synthesized from 1 μg of total RNA
using the High Capacity cDNA Reverse Transcription kit. qPCR
(Taqman method) was performed in a Sequence Detection System 7500
(Applied Biosystems Life Technologies, Foster City, CA, USA)
according to the manufacturer’s instructions. All other materials
including the Taqman dye and probes were obtained from Invitrogen.
Briefly, the samples were incubated at 25°C for 10 min for the
initial denatuation and subsequently subjected to 40 cycles of PCR,
each consisting of 37°C for 120 min and 85°C for 5 min. The β-actin
gene was used as a reference to obtain the relative fold
change.
Flow cytometry
Cells were cultured in serum-deprived α-MEM for 24 h
to induce apoptosis. Cells were harvested using trypsin/EDTA,
washed with PBS, resuspended in 1 ml binding buffer, and stained
with 10 μl Annexin V-fluorescein isothiocyanate (V-FITC) and 10 μl
propidium iodide (PI) at room temperature for 15 min. Flow
cytometry (Carl Zeiss LSM710; Carl Zeiss AG, Jena, Germany) was
used to analyze the FITC (Annexin V:FITC Apoptosis Detection kit,
BD Biosciences, San Diego, CA) and PI fluorescent (Becton,
Dickinson and Company, Franklin Lakes, NJ, USA) signals.
Caspase-3 assay
A Caspase-3 Colorimetric Assay kit (KeyGen, Nanjing,
China) was used to measure the caspase-3 activity according to the
manufacturer’s instructions. Cells were cultured in serum-deprived
α-MEM for 24 h to induce apoptosis, collected and washed with PBS.
Briefly, cells were lysed on ice in lysis buffer for 1 h and
vortexed every 20 min for 10 sec. This was followed by
centrifugation for 1 min at 12,000 × g at 4°C. Aliquots of the
supernatant containing 150 μg protein were diluted to 50 μl with
cell lysis buffer, incubated with 5 μl of substrate at 37°C for 4 h
in dark and a microplate reader (DNM-9602, Beijing Perlong Medical
Instrument Ltd) was used to measure the absorbance value of the
samples at 405 nm.
Determination of the mitochondrial DNA
(mtDNA) levels
qPCR was used to determine the relative amounts of
mtDNA. qPCR (Taqman method) was performed in the Sequence Detection
System 7500 (Applied Biosystems Life Technologies) following the
manufacturer’s instructions. Briefly, a DNA extraction kit
(Promega, Madison, WI, USA) was used to isolate the DNA from the
cells on the tenth day of differentiation. Spectrophotometry at a
wavelength of 260 nm was employed to quantify the DNA. A 110-nt
mtDNA fragment within the CYTB gene was used to quantify mtDNA. A
291-bp region of the nuclear 28S gene was used to normalize the
results. The ratio of mtDNA to nuclear DNA reflected the
concentration of mitochondria per cell.
Assessment of cellular ATP
production
On the tenth day of differentiation, a
luciferase-based luminescence assay kit (Beyotime, Nantong, China)
was used to measure the ATP content of the P19 cells.
Differentiated P19 cells were homogenized in an ice-cold
ATP-releasing buffer and a single-tube luminometer (utrao SM600;
Beyotime) was used to determine the ATP concentrations, which were
normalized to the protein concentrations.
Assessment of intracellular reactive
oxygen species (ROS) levels
A 2′,7′-dichlorodihydrofluorescein diacetate acetyl
ester (H2-DCFDA) probe (Beyotime) was used to estimate
the intracellular ROS levels. The cells were incubated with 5 μM of
H2-DCFDA for 30 min at 37°C, washed three times with
pre-warmed PBS and then observed with a confocal laser-scanning
microscope (excitation at a wavelength of 579 nm, emission at 644
nm, ×400 magnification; E4500; Nikon). Subsequently, the cells were
trypsinized and centrifuged at 1,500 × g at room temperature for 5
min, washed twice with PBS, resuspended in PBS and analyzed by flow
cytometry (Becton, Dickenson and Company).
Antibodies and western blot analysis
Cultured cells were directly transferred to tubes
containing lysis buffer and vortexed briefly. The supernatant was
collected following centrifugation at 15,200 × g for 15 min at 4°C.
Protein concentrations were determined using BCA protein assay
reagent kit (KeyGen, Nanjing, China). Total proteins were isolated
from cultured cells, separated on a 10% sodium dodecyl sulfate
(SDS) gel by SDS polyacrylamide gel electrophoresis, and
transferred onto polyvinylidene difluor-ide membranes. These
membranes were incubated with a mouse polyclonal anti-WNT1, mouse
monoclonal anti-GSK3β, rabbit polyclonal anti-β-catenin and mouse
anti-β-actin antibody (Affinity, Santa Cruz, CA, USA), and goat
anti-rabbit or rabbit anti-mouse immunoglobulin G–horseradish
peroxidase conjugate (Amersham, UK). Immunoreactive proteins were
detected by enhanced chemi-luminescence (Amersham, UK).
Luciferase assay
The recombinant vector or pGL3-Basic vector
(GenePharma, Shanghai, China) were cotransfected with the pRL-CMV
vector (GenePharma) containing a Renilla luciferase reporter
gene (as a normalizing control) into either the miR-19b knockdown
or control stable P19 cells. The Dual Luciferase Reporter Assay
system (Promega) was used to analyze the firefly and Renilla
luciferase activities 36 h later.
Statistical analysis
Each experiment was performed with at least 3
different cultures and repeated at least 3 times. Data are
presented as the mean ± standard deviation (SD). For comparison of
differences between groups, analysis of variance and unpaired
Student’s t-tests were used. P<0.05 was considered to indicate a
statistically significant difference.
Results
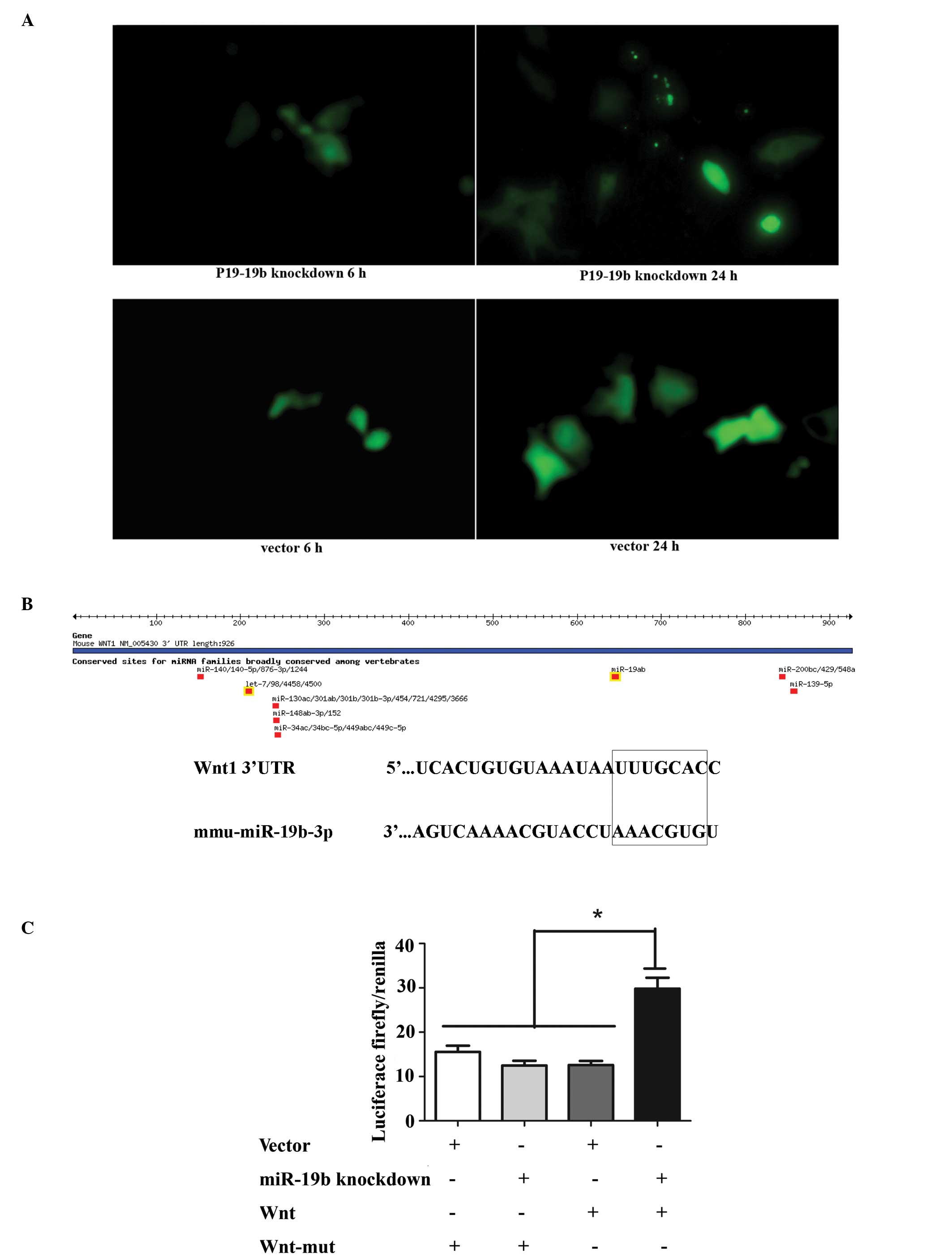
Transfection of P19 cells with the
miR-19b knockdown vector
Plasmids pGLV3/H1/eGFP/Puro-miR-19b-3p-inhibitor
sponge and pGLV3/H1/eGFP/Puro-miR-vector were transiently
transfected into P19 cells. Observation of green fluorescent
protein (GFP) expression under a fluorescence microscope indicated
similar transfection efficiencies (Fig. 1A). Subsequently, stably transfected
cells were selected by puromycin. Interaction between miRNAs and
their target site(s) in the 3′ untranslated regions (3′-UTRs)
results in translational repression or miRNA cleavage. Once the
miRNAs are inhibited, the target gene becomes free from
transcriptional repression and is activated, which can be detected
by luciferase activity. In order to knockdown miR-19b
(5′-UGUGCAAAUCCAUGCAAAACUGA-3′), complementary binding sites
(5′-TCAGTTTTGCATGGATT TGCACA-3′) were inverted into the plasmid
which were perfectly complementary with the sponge RNA (5′-GATCCT
CAGTTTTGCATGGATTTGCACACTAGTCAGTTTTGCA
TGGATTTGCACATTACCATCAGTTTTGCATGGATTTG
CACAGAATTCAGTTTTGCATGGATTTGCACATTTTTT GAATT-3′). Previous studies
have confirmed that the 3′-UTR of Wnt1 is a target of miR-19b. As
expected, miR-19b knockdown significantly rescued the luciferase
activity of the pGL3-wnt-3′-UTR reporter but not the mutated
construct (mu-pGL3-Wnt-′3-UTR) (Fig.
1B and C; P<0.01). The result of the luciferase activity
assay indirectly revealed that miR-19b was knocked down,
demonstrating that the miR-19b-knockdown vector was constructed
successfully.
miR-19b knockdown inhibits cellular
proliferation
The CCK-8 assay was used to assess the growth of
miR-19b-knockdown and control P19 cells. At days 1, 2 and 4, the
optical density (OD) values of the miR-19b-knockdown and control
cells showed no significant differences. However, at days 5, 6 and
7, the OD values of the miR-19b-knockdown cells were significantly
lower than those of the control cells. Thus, a reduced growth rate
of the miR-19b-knockdown cells was observed compared with that
observed in the control cells (Fig.
2A; P<0.05 and P<0.01). In addition, miR-19b-knockdown
also affected the cell cycle. Flow cytometry of the cell cycle
distribution detected a significantly lower percentage of
miR-19b-knockdown P19 cells in the S phase of the cell cycle
compared with that of the control cells (Fig. 2B; P<0.01).
MiR-19b knockdown reduces the levels of
apoptosis in P19 cells
Caspase-3 protein assays and Annexin V-FITC, which
binds to phosphatidylserine, were used to detect the early stages
of apoptosis. Cells were induced to undergo apoptosis via 24 h of
serum starvation. Binding experiments with Annexin V-FITC indicated
that miR-19b knockdown reduced the number of apoptotic cells in
response to serum deprivation (Fig.
3A; P<0.05). Additionally, the caspase-3 activity assay
(Fig. 3B; P<0.01) revealed that
miR-19b knockdown reduced the number of apoptotic cells in response
to serum deprivation. These results indicate that miR-19b knockdown
inhibits serum deprivation-induced apoptosis in P19 cells.
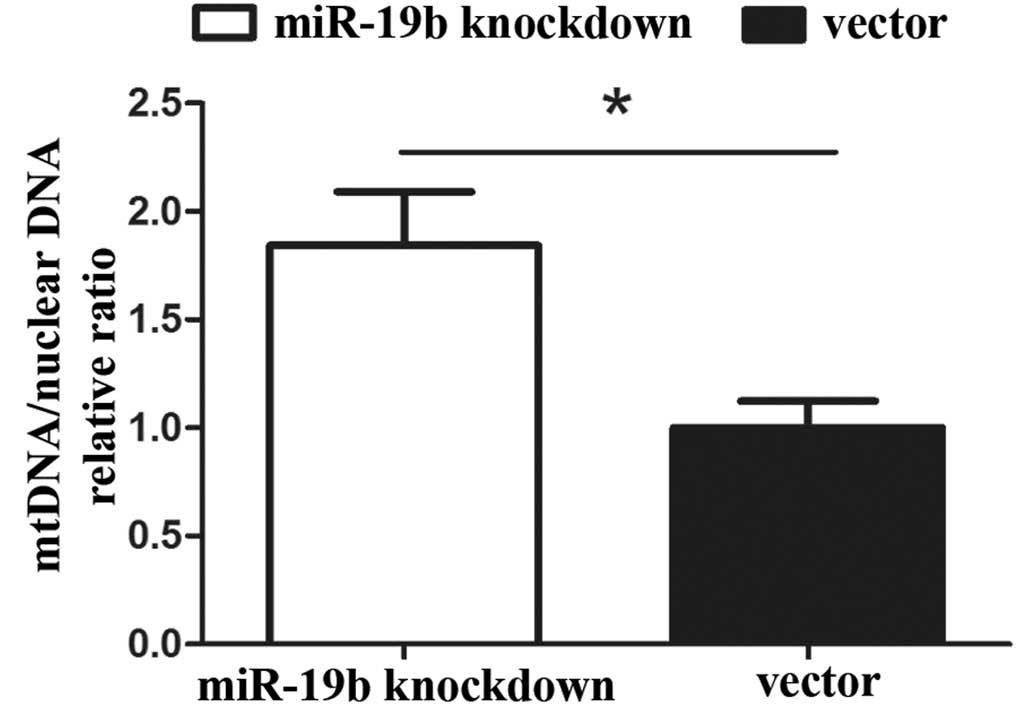
Effects of miR-19b knockdown on mtDNA
copy number in P19 cells
The relative expression levels of mitochondria were
assessed, as these represent the total mtDNA copy number in the two
cell lines. The mtDNA copy number per mitochondrion is considered
to be constant in all mammalian cell types. As a result, the copy
number of mtDNA can be determined from the relative quantity of
mitochondria. qPCR revealed that the mtDNA copy number was
significantly higher in the miR-19b knockdown group compared with
that in the vector group (Fig. 4;
P<0.05).
Cellular ATP production increases upon
miR-19b knockdown
In eukaryotic cells, the mitochondrion is the major
platform for energy transduction, producing ATP via the oxidative
metabolism of nutrients. Impaired mitochondria may lead to a
reduction in the levels of ATP. The results of the current study
revealed that the total cellular production levels of ATP were
increased in the miR-19b knockdown cells compared with those in the
control cells (Fig. 5;
P<0.01).
Effects of miR-19b knockdown on
intracellular ROS levels
Subsequently, the effect of miR-19b knockdown on the
ROS content of cells was investigated. The levels of ROS in the
miR-19b-knockdown cells were much lower than those in the control
cells (Fig. 6A), as indicated by
less intense fluorescent signals in the presence of the
H2-DCFDA (Fig. 6B).
Hence, the results clearly demonstrate that miR-19b knockdown
inhibits apoptosis, accompanied by mitochondrial dysfunction in P19
cells.
MiR-19b knockdown has no clear effect on
the morphology of P19 cells during differentiation
To investigate the ability of the miR-19b-knockdown
P19 cells to differentiate into myocardial cells, the morphological
appearance was observed and myocardial-specific molecular markers
(cTnT, GATA4 and NKX2.5) were quantified, and the appearance of
beating cell clusters during DMSO-induced differentiation was
monitored (Fig. 7A). However, no
differences were observed in the cell morphology or the time taken
for the appearance of beating cell clusters between the
miR-19b-knockdown cells and the control cells. In addition to
observing cell morphology, the expression of myocardial-specific
molecular markers was analyzed at the RNA level, including cTnI,
GATA4 and NKX2.5, which are known to have upregulated levels of
expression during the differentiation of mouse P19 cells into
myocardial cells (Fig. 7B;
P<0.05). The expression levels of these marker genes were
detected by RT-qPCR in the two cell lines on days 0, 4, 8, 10 and
12. Expression levels of all the marker genes gradually increased
during the process of differentiation, however, in the
miR-19b-knockdown cells only cTnT showed significantly lower
expression levels compared with those observed in the control cells
at days 4, 10 and 12.
Effect of miR-19b knockdown on the
Wnt/β-catenin signaling pathway
Wnt is an essential regulator of cell
differentiation. The Wnt protein initiates the Wnt/β-catenin
signaling pathway, GSK3β acts as a switch and β-catenin functions
as the effector molecule. Therefore, qPCR and western blot analyses
were used to detect the RNA and protein expression changes of
several key molecules in the Wnt signaling pathway (Wnt, GSK3β and
β-catenin). During the induction of differentiation of P19 cells
into myocardial cells, the relative expression levels of Wnt, GSK3β
and β-catenin were significantly higher compared with those in the
vector group at almost all time points (Fig. 8A; P<0.05 and P<0.01). At the
protein level, the expression levels of Wnt in the
miR-19b-knockdown group were significantly higher than those in the
vector group at days 0, 10 and 12, those of β-catenin were
significantly higher than those in the vector group at days 8 and
12, those of GSK3β were significantly higher than those in the
vector group at day 4, and the trend in the levels of β-catenin
expression was similar to that of Wnt1 (Fig. 8B and C, P<0.05).
Discussion
MiRNAs contribute to cardiac development and can be
used as novel biomarkers and therapeutic targets for CHDs (2,14).
Previous studies have determined that the miR-17–92 cluster, which
includes mir-19b, is important in a number of diseases and miR-19b
expression may correlate with the incidence of cardiovascular
diseases and cardiogenesis (15).
Gao et al (16) found that
the downregulation of miR-19b contributes to angiotensin II-induced
overexpression of connective tissue growth factor in
cardiomyocytes. Jung et al (17) determined that there is an
interaction between the hepatitis B virus (HBV) and the miR-17–92
polycistron via c-Myc, and that miR-20a and miR-92a-1 induce
post-transcriptional suppression of HBV. In the present study, the
P19 cell line was used as a research model, and a stable line of
miR-19b-knockdown P19 cells was established to evaluate the effect
of this miRNA on P19 cells and their differentiation toward
myocardial cells. MiR-19b knockdown inhibited proliferation and
apoptosis in the P19 cells. Notably, a previous study found that
the overexpression of miR-19b could increase proliferation, inhibit
apoptosis and promote differentiation of P19 cells into mature
cardiac cells (18). This
indicates that overexpression of miR-19b or miR-19b knockdown may
influence morphogenesis in the embryonic heart by inhibiting
excessive apoptosis in the myocardium. Embryonic fetal heart growth
depends on the balance between cardiomyocyte proliferation and
apoptosis (19). Inadequate
proliferation or excess apoptosis may directly or indirectly result
in CHD (20), which is often
caused by altered proliferation and/or apoptosis in the septum,
neighboring tissue or myocardium (21).
The results of the present study, including those
from the CCK-8 assay and cell cycle analysis, indicate that miR-19b
knockdown inhibits the proliferation of P19 cells by significantly
reducing the percentage of cells in the S phase, however, the
specific mechanism by which this occurs remains to be determined.
Yan et al (22) determined
that overexpression of the miR-17–92 cluster markedly inhibited
hypoxia-induced apoptosis. Sharifi et al (23) found that inhibition of miR-92a
inhibited cell proliferation in human acute promyelocytic leukemia.
In the current study, miR-19b knockdown was determined to
significantly inhibit serum starvation-induced apoptosis. The
molecular mechanism that underlies this effect remains unknown.
Crow et al (24)
hypothesized that mitochondria are important in the transmission
and amplification of apoptotic signals. Mitochondria are at the
center of the regulatory processes for apoptosis (25). In the present study, it was
demonstrated that levels of intracellular ROS were reduced, ATP
contents were increased and the levels of mitochondrial DNA were
increased on the tenth day of differentiation of P19 cells. It may
be hypothesized that miR-19b knockdown cells generate more ATP to
compensate for lost ATP production, which correlates with the
increased level of mitochondrial DNA. It was observed that miR-19b
knockdown does not significantly affect the differentiation of P19
cells into cardiomyocytes, indicated by the lack of morphological
changes and the normal expression of cardiomyogenesis-specific
molecular markers. Although the expression of all the marker genes
(cTnT, GATA4 and NKX2.5) gradually increased during
differentiation, only cTnT in miR-19b-knockdown cells showed
significantly lower expression levels than those in the control
cells at days 4, 10 and 12.
Furthermore, the results of the present study
demonstrate that miR-19b does affect the Wnt/β-catenin signaling
pathway. Wnt signaling is an essential regulator of cardiovascular
differentiation, morphogenesis and progenitor self-renewal
(26). Given that miRNAs
negatively regulate their targets, miR-19b knockdown should
upregulate its potential targets. A previous study revealed that
miR-19b may indirectly target Wnt1 mRNA through its 3′-UTR
(16). In the present study, the
results of the luciferase assay indicate that miR-19b knockdown
rescued Wnt1 expression by removing its interaction with its
cognate miRNA. Wnts were initially considered suppressive of heart
formation. Wnts 1, 3A and 8 act via the inhibition of GSK3,
allowing nuclear localization of β-catenin, which appears to
inhibit cardiac differentiation, whereas the non-canonical Wnt11,
along with protein kinase C, appears to enhance cardiac
differentiation (27). Previous
results in zebrafish indicate that β-catenin signaling is blocked
in heart valve formation, which demonstrates a negative role of
Wnts in heart development (28).
In the present study, miR-19b knockdown affected differentiation by
increasing the activation of the Wnt/β-catenin signaling pathway
via an essential upstream target of Wnt1.
Furthermore, Wnt/β-catenin signaling can promote
cardiogenesis by inducing the proliferation of cardiac progenitor
cells in the secondary heart field (29–31).
Wnt/β-catenin signaling is also important in the endocardium, where
it regulates the specification and proliferation of endocardial
cushion cells (32). In the
present study, following the successful knockdown of miR-19b, which
normally acts on the 3′-UTR of Wnt1, the levels of Wnt1 protein
expression significantly increased, thereby activating
Wnt/β-catenin signaling and inhibiting myocardial cell
development.
Previous studies indicate that endogenous miR-19b
may have a key regulatory role in constraining the production of
pro-inflammatory cytokines and chemokines by fibroblast-like
synoviocytes and hence contribute to the pathology of inflammation
(33). MiR-19b is also a novel
regulator of fibrotic TGF-β signaling and the loss of miR-19b
following hepatic stellate cell (HSC) activation perpetuates the
fibrotic response (34).
Furthermore, the miR-19a/b family regulates cardiac hypertrophy and
survival by repressing the target genes atrogin-1 and MuRF-1
(35).
In conclusion, the results of the present study
demonstrate that miR-19b knockdown significantly inhibits the
proliferation and apoptosis of P19 cells. MiR-19b knockdown results
in an increase in Wnt expression levels, which activates the
Wnt/β-catenin signaling pathway in P19 cells, and may regulate the
cardiomyocyte differentiation of P19 cells. These results indicate
that miR-19b overexpression and knockdown leads to an imbalance
between proliferation and apoptosis, which may result in embryonic
cardiac malformations. This study of miR-19b provides insight into
novel therapeutic strategies for CHD. Further study into the
functions of related miRNAs may elucidate the processes of
pathogenesis during cardiogenesis. However, the molecular
mechanisms that mediate the balance between proliferation and
apoptosis in response to miR-19b knockdown or overexpression
require further investigation.
Acknowledgements
This study was supported by grants from the National
Natural Science Foundation of China (grant no. 81070500), the Key
Medical Personnel Foundation of Jiangsu Province (grant no.
RC2011021), the Nanjing Medical Science and Technique Development
Foundation (grant no. QRX11107) and the Science and Technology
Development Foundation of Nanjing Medical University (grant no.
2010NJMUZ15).
References
|
1
|
Capozzi G, Caputo S, Pizzuti R, Martina L,
Santoro M, Santoro G, et al: Congenital heart disease in live-born
children: incidence, distribution, and yearly changes in the
Campania Region. J Cardiovasc Med (Hagerstown). 9:368–374. 2008.
View Article : Google Scholar
|
|
2
|
Thum T, Catalucci D and Bauersachs J:
MicroRNAs: novel regulators in cardiac development and disease.
Cardiovasc Res. 79:562–570. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wu C, Cao Y, He Z, He J, Hu C, Duan H and
Jiang J: Serum levels of miR-19b and miR-146a as prognostic
biomarkers for non-small cell lung cancer. Tohoku J Exp Med.
232:85–95. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lu Y, Thomson JM, Wong HY, Hammond SM and
Hogan BL: Transgenic over-expression of the microRNA miR-17–92
cluster promotes proliferation and inhibits differentiation of lung
epithelial progenitor cells. Dev Biol. 310:442–453. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Boggs RM, Moody JA, Long CR, Tsai KL and
Murphy KE: Identification, amplification and characterization of
miR-17–92 from canine tissue. Gene. 404:25–30. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hackl M, Brunner S, Fortschegger K,
Schreiner C, Micutkova L, Mück C, Laschober GT, et al: miR-17,
miR-19b, miR-20a, and miR-106a are down-regulated in human aging.
Aging Cell. 9:291–296. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Skerjanc IS: Cardiac and skeletal muscle
development in P19 embryonal carcinoma cells. Trends Cardiovasc
Med. 9:139–143. 1999. View Article : Google Scholar
|
|
8
|
van der Heyden MA and Defize LH: Twenty
one years of P19 cells: what an embryonal carcinoma cell line
taught us about cardiomyocyte differentiation. Cardiovasc Res.
58:292–302. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
van der Heyden MA, van Kempen MJ, Tsuji Y,
Rook MB, Jongsma HJ and Opthof T: P19 embryonal carcinoma cells: a
suitable model system for cardiac electrophysiological
differentiation at the molecular and functional level. Cardiovasc
Res. 58:410–422. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Han SP, Pan Y, Peng YZ, Gu XQ, Chen RH and
Guo XR: Folbp1 promotes embryonic myocardial cell proliferation and
apoptosis through the WNT signal transduction pathway. Int J Mol
Med. 23:321–330. 2009.PubMed/NCBI
|
|
11
|
Hu DL, Chen FK, Liu YQ, Shen YH, Yang R,
et al: GATA-4 promotes the differentiation of P19 cells into
cardiac myocytes. Int J Mol Med. 26:365–372. 2010.PubMed/NCBI
|
|
12
|
Cohen ED, Tian Y and Morrisey EE: Wnt
signaling: an essential regulator of cardiovascular
differentiation, morphogenesis and progenitor self-renewal.
Development. 135:789–798. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ueno S, Weidinger G, Osugi T, Kohn AD,
Golob JL, et al: Biphasic role for Wnt/beta-catenin signaling in
cardiac specification in zebrafish and embryonic stem cells. Proc
Natl Acad Sci USA. 104:9685–9690. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhu S, Cao L, Zhu J, Kong L, Jin J, Qian
L, Zhu C, Hu X, Li M, Guo X, Han S and Yu Z: Identification of
maternal serum microRNAs as novel non-invasive biomarkers for
prenatal detection of fetal congenital heart defects. Clin Chim
Acta. 424:66–72. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
van Almen GC, Verhesen W, van Leeuwen RE,
van de Vrie M, Eurlings C, et al: MicroRNA-18 and microRNA-19
regulate CTGF and TSP-1 expression in age-related heart failure.
Aging Cell. 10:769–779. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Gao S, Liu TW, Wang Z, Jiao ZY, Cai J, Chi
HJ and Yang XC: Downregulation of microRNA-19b contributes to
angiotensin II-induced overexpression of connective tissue growth
factor in cardiomyocytes. Cardiology. 127:114–120. 2014. View Article : Google Scholar
|
|
17
|
Jung YJ, Kim JW, Park SJ, Min BY, Jang ES,
Kim NY, Jeong SH, Shin CM, Lee SH, Park YS, Hwang JH, Kim N and Lee
DH: c-Myc-mediated overexpression of miR-17–92 suppresses
replication of hepatitis B virus in human hepatoma cells. J Med
Virol. 85:969–978. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Qin DN, Qian L, Hu DL, Yu ZB, Han SP, Zhu
C, Wang X and Hu X: Effects of miR-19b overexpression on
proliferation, differentiation, apoptosis and Wnt/β-catenin
signaling pathway in P19 cell model of cardiac differentiation in
vitro. Cell Biochem Biophys. 66:709–722. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Fiorina P, Corradi D, Pinelli S, Maestri
R, Lagrasta C, Buscaglia M, et al: Apoptotic/mytogenic pathways
during human heart development. Int J Cardiol. 96:409–417. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lévy M, Maurey C, Celermajer DS, Vouhé PR,
Danel C, Bonnet D and Israël-Biet D: Impaired apoptosis of
pulmonary endothelial cells is associated with intimal
proliferation and irreversibility of pulmonary hypertension in
congenital heart disease. J Am Coll Cardiol. 49:803–810. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gittenberger-de Groot GA, Bartelings MM,
Deruiter MC and Poelmann RE: Basics of cardiac development for the
understanding of congenital heart malformations. Pediatr Res.
57:169–176. 2005. View Article : Google Scholar
|
|
22
|
Yan HL, Xue G, Mei Q, Wang YZ, Ding FX,
Liu MF, et al: Repression of the miR-17–92 cluster by p53 has an
important function in hypoxia-induced apoptosis. EMBO J.
28:2719–2732. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Sharifi M, Salehi R, Gheisari Y and Kazemi
M: Inhibition of MicroRNA miR-92a inhibits cell proliferation in
human acute promyelocytic leukemia. Turk J Hematol. 30:157–162.
2013. View Article : Google Scholar
|
|
24
|
Crow MT, Mani K, Nam YJ and Kitsis RN: The
mitochondrial death pathway and cardiac myocyte apoptosis. Circ
Res. 95:957–970. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Brooks C and Dong Z: Regulation of
mitochondrial morphological dynamics during apoptosis by Bcl-2
family proteins: a key in Bak? Cell Cycle. 6:3043–3047. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Cohen ED, Tian Y and Morrisey EE: Wnt
signaling: an essential regulator of cardiovascular
differentiation, morphogenesis and progenitor self-renewal.
Development. 135:789–798. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Olson EN and Schneider MD: Sizing up the
heart: development redux in disease. Genes Dev. 17:1937–1956. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hurlstone AF, Haramis AP, Wienholds E,
Begthel H, Korving J, Van Eeden F, et al: The Wnt/beta-catenin
pathway regulates cardiac valve formation. Nature. 425:633–637.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Verhoeven MC, Haase C, Christoffels VM,
Weidinger G and Bakkers J: Wnt Signaling Regulates Atrioventricular
Canal Formation Upstream of BMP and Tbx2. Birth Defect Res A Clin
Mol Teratol. 91:435–440. 2011. View Article : Google Scholar
|
|
30
|
Ai D, Fu X, Wang J, Lu MF, Chen L, Baldini
A, et al: Canonical Wnt signaling functions in second heart field
to promote right ventricular growth. Proc Natl Acad Sci USA.
104:9319–9324. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kwon C, Arnold J, Hsiao EC, Taketo MM,
Conklin BR and Srivastava D: Canonical Wnt signaling is a positive
regulator of mammalian cardiac progenitors. Proc Natl Acad Sci USA.
104:10894–10899. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Cai X, Zhang W, Hu J, Zhang L, Sultana N,
Wu B, Cai W, Zhou B and Cai CL: Tbx20 acts upstream of Wnt
signaling to regulate endocardial cushion formation and valve
remodeling during mouse cardiogenesis. Development. 140:3176–3187.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Gantier MP, Stunden HJ, McCoy CE, Behlke
MA, Wang D, Kaparakis-Liaskos M, Sarvestani ST, Yang YH, Xu D, Corr
SC, Morand EF and Williams BR: A miR-19 regulon that controls NF-iB
signaling. Nucleic Acids Res. 40:8048–8058. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Lakner AM, Steuerwald NM, Walling TL,
Ghosh S, Li T, McKillop IH, Russo MW, Bonkovsky HL and Schrum LW:
Inhibitory effects of microRNA 19b in hepatic stellate
cell-mediated fibrogenesis. Hepatology. 56:300–310. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Song DW, Ryu JY, Kim JO, Kwon EJ and Kim
do H: The miR-19a/bfamily positively regulates cardiomyocyte
hypertrophy by targeting atrogin-1 and MuRF-1. Biochem J.
457:151–162. 2014. View Article : Google Scholar
|