Introduction
Aquaporins (AQPs) are a family of ubiquitous
membrane proteins that form pores for the selective permeation of
water and other small molecules (1). Aquaglyceroporins belong to a subgroup
of the AQP family and are able to transport small organic
compounds, such as glycerol or urea. Overall, five molecules (AQP3,
AQP7, AQP9, AQP10 and bacterial glycerol facilitator) have been
classified as aquaglyceroporins (2). AQP9 is most abundantly expressed in
the liver (3). Rojek et al
(4) reported that AQP9 knockout
mice exhibit hypertriacylglycerolemia, a sign of metabolic
syndrome. AQP9 is implicated in hepatic glycerol transport and
consequently contributes to neoglucogenesis (5). Therefore, the dysregulation of AQP9
gene expression is important in the pathogenesis of metabolic
disorders.
Compelling evidence has indicated that insulin acts
as a key regulator of AQP9 (6,7). The
AQP9 promoter contains a negative insulin response element
(IRE), TGTTTTC, at −496/−502 and AQP9 mRNA expression is
downregulated by insulin in cultured hepatocytes (6). Rodríguez et al (8) observed that insulin inhibited the
expression of AQP9 via the PI3K/Akt/mTOR signaling pathway. In a
rat model, hepatic AQP9 expression levels were found to fluctuate
with circulating insulin levels (9). Forkhead box protein O1 (FOXO1) is a
forkhead transcriptional factor that mediates the regulatory
effects of insulin on target gene expression (10,11).
Tsuchida et al (12)
reported that insulin negatively modulates the expression levels of
adiponectin receptors via the PI3K/FOXO1-dependent pathway. A
previous study demonstrated that FOXO1 was implicated in the
regulation of AQP9 expression, as depletion of FOXO1 using small
interfering RNA technology was observed to reduce the
transcriptional activation of AQP9 (13). These findings suggest the
involvement of insulin/FOXO1 signaling in the regulation of AQP9
expression.
Chromatin consists of repeating units of
nucleosomes, which consist of ~146 bp DNA wrapped around an octamer
of four core histone proteins (H3, H4, H2A and H2B) (14). Chromatin remodeling is pivotal in
regulating gene expression, which modulates the accessibility of
genomic DNA to regulatory transcription machinery proteins.
Covalent modifications are important mechanisms contributing to
such remodeling, and include the acetylation, phosphorylation and
methylation of histone proteins in the nucleosome (15). Insulin has been shown to alter
chromatin structure via the promotion of histone H3
post-translational modifications (16,17).
FOXO1 has the ability to initiate and dynamically modulate active
chromatin states (18). Therefore,
insulin and FOXO1 were hypothesized to regulate AQP9 expression in
part through an epigenetic mechanism involving post-translational
modifications of histone H3. To analyze this hypothesis, in the
present study, the effects of insulin treatment and FOXO1
overexpression on AQP9 expression levels, and the histone H3
modifications at the AQP9 gene promoter in HepG2 human hepatocytes,
were examined.
Materials and methods
Cells and reagents
The cells and reagents used in this study were as
follows: HepG2 and Hek 293T human cells (Type Culture Collection of
Chinese Academy of Sciences, Shanghai, China), fetal bovine serum
(FBS; Gibco-BRL, Carlsbad, CA, USA), Dulbecco’s modified Eagle’s
medium (DMEM; Hyclone Laboratories, Inc., Logan, UT, USA), reagents
for quantitative polymerase chain reaction (qPCR; Takara
Biotechnology Dalian Co., Ltd., Dalian, China), Lipofectamine™ 2000
(Invitrogen Life Technologies, Carlsbad, CA, USA), and polybrene
and recombinant human insulin (Sigma-Aldrich, St. Louis, MO, USA).
Primary antibodies against FOXO1 (Abcam, Cambridge, UK), AQP9
(Santa Cruz Biotechnology, Santa Cruz, CA, USA) and β-actin
(Beijing Zhongshan Golden Bridge Biotechnology Co., Ltd., Beijing,
China) were used for western blotting.
Cell culture and treatment
The HepG2 and 293T cells were cultured in DMEM
supplemented with 10% FBS, 100 U/ml penicillin and 100 μg/ml
streptomycin at 37°C in a humidified 5% CO2 atmosphere.
For insulin treatment, HepG2 cells were seeded at a density of
5×104 cells/ml into 6-well plates and incubated in
complete medium with 10% FBS for 24 h. At ~50% confluence, the
cells were starved in FBS-free medium for 6 h followed by
stimulation with 500 μM insulin for different durations. Following
the treatment, the cells were harvested for further analysis.
Plasmid construction
Total RNA was isolated from the HEK293T cells using
TRIzol™ (Invitrogen Life Technologies) according to the
manufacturer’s instructions and then reverse-transcribed to cDNA.
The cDNA sequence encoding full-length human FOXO1 (GenBank no.
NM_002015.3) was amplified by PCR. The PCR primers were as follows:
Forward, 5′-AAAGCTAGCATGGCCGAGGCGCCTCAG-3′ and reverse,
5′-AAAACTAGTTCAGCCTGACACCCAGCTA-3′. The PCR product was cloned into
a pWPI vector (Addgene, Cambridge, MA, USA) and the sequence was
confirmed by DNA sequencing.
Preparation and transduction of
lentiviral particles
For the production of lentiviral particles
expressing FOXO1, HEK293T cells were transfected with the
expression vector WPI-FOXO1 along with the packaging vectors psPAX2
and pMD (Addgene) using Lipofectamine 2000 (Invitrogen Life
Technologies), according to the manufacturer’s instructions.
Following incubation for 48 h, the medium containing the lentiviral
particles was collected and centrifuged, and aliquots were stored
at −80°C until further use.
For lentiviral particle transduction, the HepG2
cells were seeded in 60 mm dishes and 3 ml viral supernatant was
added to 2 ml DMEM with 10% FBS following cell attachment. The
cells were infected for 24 h in the presence of polybrene (8
μg/ml). After 24 h, the cell culture medium was refreshed. Since
the pWPI vector expresses green fluorescent protein (GFP), the
transfection efficiency was monitored by detecting GFP expression
levels by fluorescence microscopy.
Reverse transcription and qPCR
Total RNA was isolated from the HepG2 cells
following treatment using TRIzol and cDNA was reverse-transcribed
from 1 μg total RNA sample. qPCR was performed using a SYBR Green
PCR Master Mix kit (Takara Biotechnology Dalian Co., Ltd Dalian,
China). The PCR primers were as follows: AQP9 forward,
5′-CTCCTGATTATTGTCATTGC-3′ and AQP9 reverse,
5′-ATCCACCAGAAGTTGTTT-3′; β-actin forward, 5′-CCTGGCACCCAGCACAAT-3′
and β-actin reverse, 5′-GCCGATCCACACGGAGTA-3′. The cycling
conditions were as follows: Initial denaturation at 95°C for 3 min,
40 cycles of denaturation at 95°C for 10 sec and annealing at 60°C
for 30 sec. The data were analyzed with CFX96 Manager software
(Bio-Rad Laboratories, Munich, Germany). The relative AQP9 mRNA
levels were calculated following normalization to β-actin mRNA
levels.
Western blot analysis
Subsequent to treatment, the HepG2 cells were lysed
in radioimmunoprecipitation assay buffer containing 25 mM Tris-HCl
(pH 8.0), 1% Nonidet-P-40, 0.5% sodium deoxycholate, 0.1% sodium
dodecyl sulfate, 125 mM NaCl and 1% phenylmethanesulfonyl fluoride
(Sigma-Aldrich) for 30 min at 4°C. The total protein was measured
using a Bicinchoninic Protein Assay kit (Pierce Biotechnology,
Inc., Rockford, IL, USA). Samples of the total protein extracts
(~60 μg) were separated by 12% SDS-PAGE and transferred to a
polyvinylidene fluoride membrane. The membrane was incubated
overnight with primary antibodies at 4°C (anti-AQP9, 1:20 dilution;
anti-FOXO1, 1:1,000 dilution). Subsequent to washing three times,
the membrane was incubated for 1 h with horseradish
peroxidase-conjugated secondary antibodies (1:3,000 dilution;
Beijing Zhongshan Golden Bridge Biotechnology Co., Ltd.). The
protein expression was visualized using an Enhanced
Chemiluminescence Detection kit (Amersham Pharmacia Biotech,
Amersham, UK). The relative intensities of the bands were
determined by densitometry using Quantity One software (Bio-Rad,
Hercules, CA, USA).
Chromatin immunoprecipitation (ChIP)-PCR
assay
ChIP experiments were performed using a Magna ChIP A
Chromatin Immunoprecipitation kit (Milipore, Billerica, MA, USA)
according to the manufacturer’s instructions. Briefly, the cells
were incubated in 1% formaldehyde at room temperature for 10 min,
followed by incubation for 10 min in ice-cold lysis buffer
containing a mixture of protease inhibitors. The cells were then
sonicated 10 times at 1 min intervals. Sonication yielded DNA
fragments ~250 bp in length, as determined by agarose gel
electrophoresis. Following centrifugation, the sonicated sample was
diluted 1:10 with dilution buffer, and 20 μl diluted supernatant
served as an input control. The chromatin solution was precleared
using Salmon Sperm DNA/Protein A Agarose Slurry (Millipore,
Bedford, MA, USA) for 30 min and incubated overnight at 4°C with
anti-acetylated histone H3. The immunoprecipitated complexes were
recovered by adding 30 μl Salmon Sperm DNA/Protein A Agarose
Slurry. Following washing, 5 M NaCl was added to reverse the
formaldehyde cross-linking and the pellets were treated with
proteinase K. DNA samples were purified using the QIAquick PCR
purification kit (Qiagen, Valencia, CA, USA). Immunoprecipitated
DNA and input DNA were amplified by qPCR using the SYBR Green PCR
Master Mix kit (Takara Biotechnology Dalian Co., Ltd). The primers
used to amplify the IRE locus of the AQP9 promoter were as
follows: Forward, 5′-ATTTCGGGTTCTAAGTCGC-3′ and reverse,
5′-TTCCTGGAGATGTCTGGTAAG-3′. All assays were performed in
triplicate. The percentage enrichment of immunoprecipitated DNA was
calculated relative to the input DNA. The ChIP results were
normalized to the input DNA and are expressed as the fold
enrichment relative to untreated cells (assigned 1-fold).
Statistical analysis
Statistical analyses were conducted using SPSS
version 18.0 (SPSS, Inc., Chicago, IL, USA). The results are
presented as the means ± standard deviation. Significance was
determined by Student’s t-test or one-way analysis of variance with
a Student-Newman-Keuls post hoc test. A P<0.05 was
considered to indicate a statistically significant difference.
Results
Insulin suppresses AQP9 mRNA expression
in a time-dependent manner
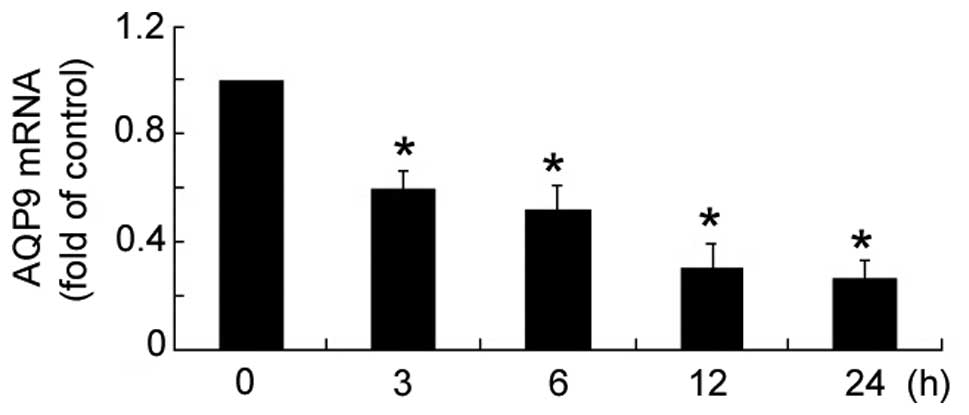
Insulin treatment resulted in a significant
(P<0.05) reduction in AQP9 mRNA expression levels in HepG2
cells, as compared with untreated cells (Fig. 1). Furthermore, the reduction
occurred in a time-dependent manner, with maximum reduction
observed at 12 h.
Effects of insulin on the
post-transcriptional modifications of histone H3 at the AQP9
promoter
To verify whether insulin-mediated repression of
AQP9 expression was associated with alterations in chromatin
remodeling, ChIP and PCR analysis was performed to examine the
post-translational modifications of histone H3 at the AQP9
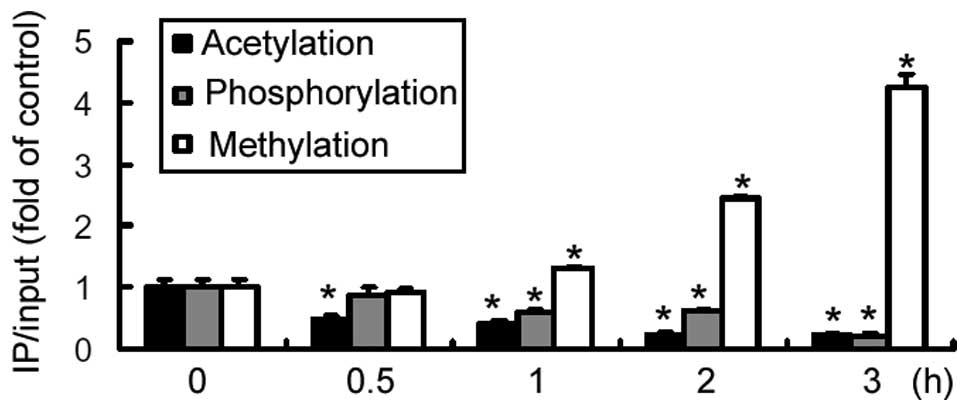
promoter. The results revealed that treatment with insulin resulted
in a marked reduction in the acetylation and phosphorylation of
histone H3 at the IRE of the AQP9 promoter, initiated at 0.5
h and 1 h, respectively, and peaking at 3 h treatment (Fig. 2). By contrast, a significant
(P<0.05) increase was detected in histone H3 methylation at the
IRE locus of the AQP9 promoter upon exposure to insulin
(Fig. 2). However, no evident
change in histone H3 modification was observed at a control site
within the second exon of AQP9 (data not shown). These
results indicate that repression of AQP9 expression by insulin is
associated with epigenetic modifications at the promoter.
Enforced expression of FOXO1 stimulates
AQP9 expression in HepG2 cells
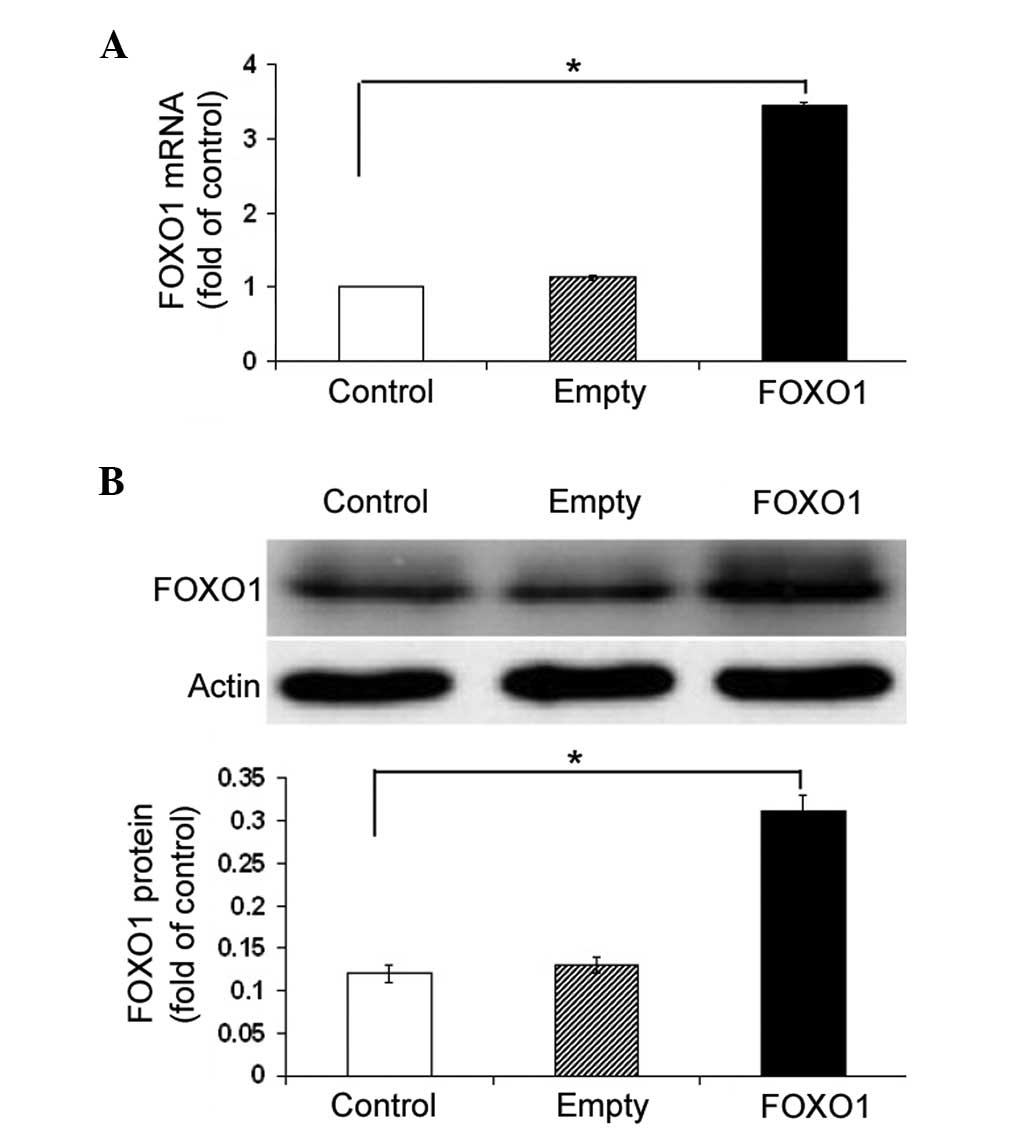
The effect of FOXO1 overexpression on AQP9
expression levels was examined. FOXO1-overexpressing plasmid
transfection resulted in a ~three-fold increase in the FOXO1 mRNA
and protein expression levels in HepG2 cells, as compared with
those of the control cells (P<0.05; Fig. 3A and B). Notably, ectopic
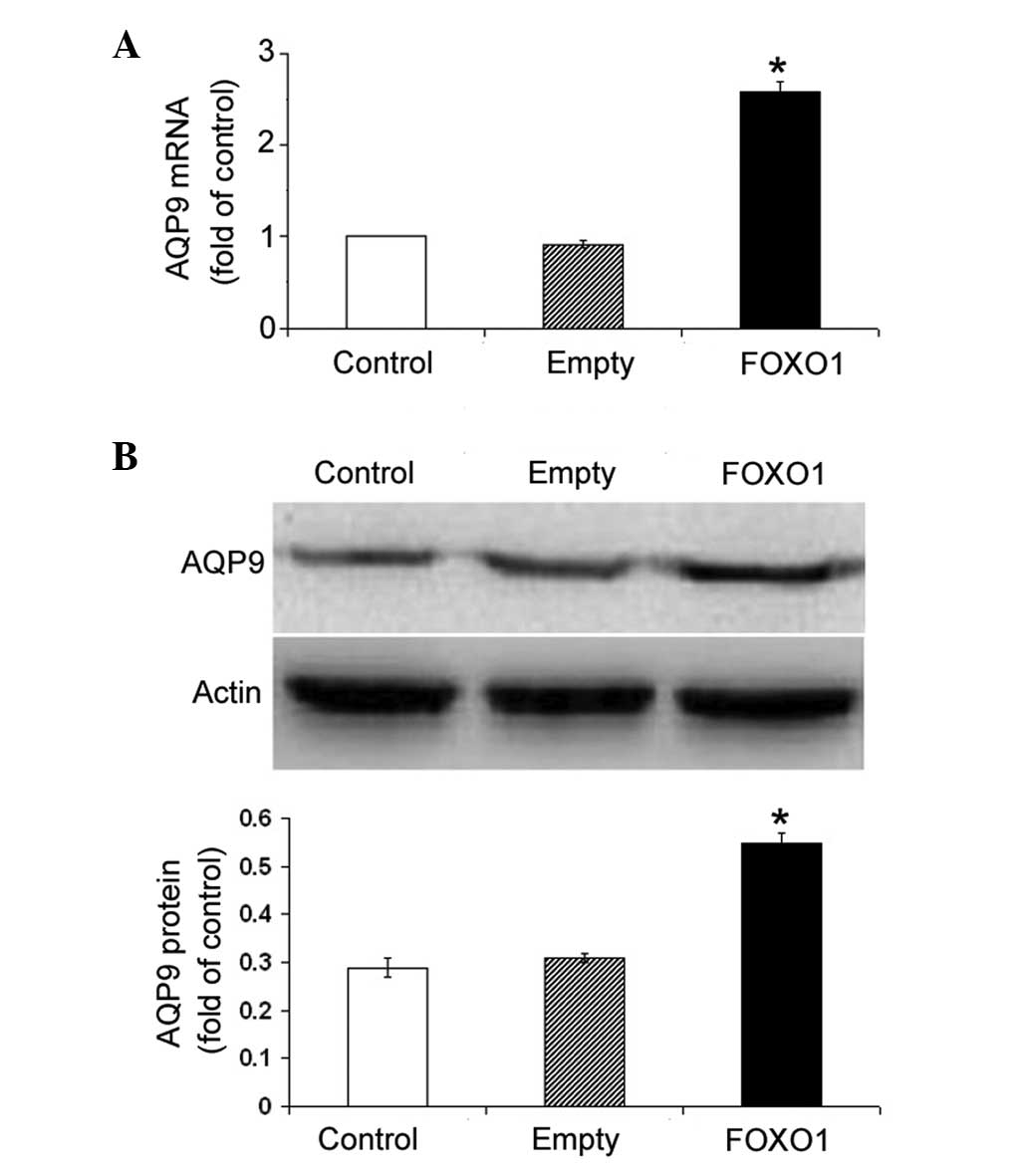
expression of FOXO1 significantly (P<0.05) increased the
abundance of AQP9 mRNA in HepG2 cells, as compared with the
non-transfected cells (Fig. 4A).
The induction of AQP9 by FOXO1 overexpression was further confirmed
at the protein level by western blotting (P<0.05; Fig. 4B).
FOXO1 promotes post-transcriptional
modifications of histone H3 at the AQP9 promoter
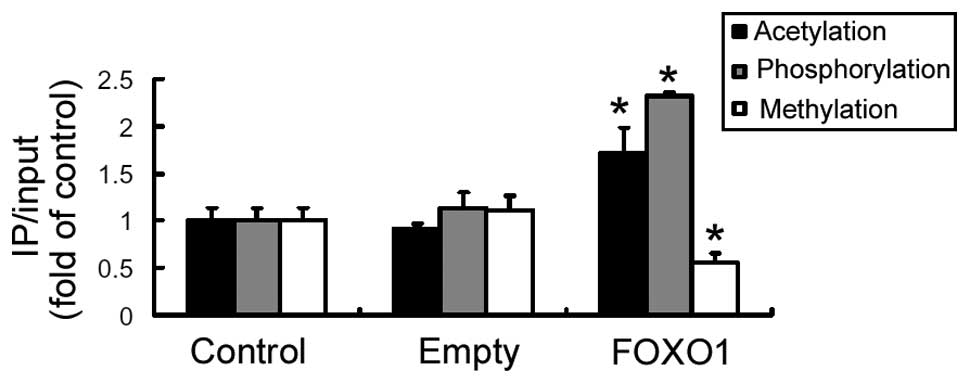
The impact of FOXO1 on epigenetic modifications of
the AQP9 promoter was analyzed. As shown in Fig. 5, the levels of histone H3
acetylation and phosphorylation at the IRE of the AQP9
promoter was significantly (P<0.05) elevated in the
FOXO1-overexpressing cells, as compared with the control cells. By
contrast, the level of histone H3 methylation at this locus was
significantly reduced by FOXO1 overexpression (P<0.05; Fig. 5).
Discussion
Glycerol is a predominant substrate in hepatic
gluconeogenesis, and the efflux of lipolytic glycerol between
adipocytes and the liver is important in modulating lipid and
glucose homeostasis. The liver-specific expression of AQP9
facilitates glycerol influx into hepatocytes, and dysregulation of
AQP9 is associated with the development of metabolic syndrome
(3). Numerous studies have shown
that AQP9 expression is negatively regulated in response to insulin
(6–8). The data from the present study
confirmed the suppression of AQP9 expression in hepatocytes by
insulin. Furthermore, this suppression occurred in a time-dependent
manner, initiated at 3 h and reaching a peak after 24 h treatment.
Kuriyama et al (6)
identified a consensus IRE in the AQP9 gene promoter, which
may be relevant to the downregulation of AQP9 by insulin.
Similarly, insulin has been found to regulate the expression of
various target genes via IRE-dependent mechanisms (19,20).
Ge et al (20) reported
that insulin stimulates the transcription of human acyl-coenzyme
A:cholesterol acyltransferase 1 (ACAT1) through an interaction of
the functional IRE upstream of the ACAT1 P1 promoter with the
CCAAT/enhancer-binding protein α (C/EBPα).
The present study provides, to the best of our
knowledge, the first evidence that insulin alters the
post-transcriptional modifications of histone H3 at the IRE locus
of the AQP9 promoter, which signifies another mechanism of
downregulation of AQP9 expression by insulin. Numerous reversible
histone covalent modifications, including acetylation,
phosphorylation and methylation, have been associated with distinct
transcription states (21).
Histone H3 hyperacetylation is commonly associated with the
alleviation of repressive histone-DNA interactions, facilitating
the transcription process. By contrast, H3 methylation at lysine 9
is generally associated with the assembly of compact or closed
chromatin surrounding the DNA, resulting in gene silencing. These
histone modifications, individually and together, can modulate
chromatin structure and gene expression (22). Cheung et al (23) reported that epidermal growth
factor-induced H3 phosphorylation affects subsequent acetylation
reactions in mammalian cells. The results from the present study
demonstrated that the levels of H3 acetylation and phosphorylation
at the IRE locus were significantly reduced 0.5 and 1 h after
insulin treatment, respectively. Similar to H3 phosphorylation,
insulin-induced H3 methylation occurred 1 h after treatment. These
results suggest that insulin treatment resulted in sequential
deacetylation and dephosphorylation/methylation of H3, which may
cooperatively establish a repressive chromatin configuration.
Numerous transcription factors, such as C/EBPα,
specificity protein 1, activator protein 1 and FOXO1, have been
shown to mediate target gene expression following the
administration of insulin (19,20).
The present study revealed that the enforced expression of FOXO1
resulted in a significant elevation in AQP9 expression levels in
HepG2 cells. Furthermore, FOXO1 overexpression induced H3
acetylation and phosphorylation, and reduced H3 methylation in the
IRE locus of the AQP9 promoter, suggesting the formation of
a permissive chromatin structure. These results are contrary to the
effects of insulin on AQP9 expression and local histone
modifications. In addition, the induction of active chromatin
states by FOXO1 has been described in a previous study (18). Insulin has the ability to
negatively regulate the expression and transcriptional activity of
FOXO1 (24,25). FOXO1 has been reported to confer an
inhibitory effect of insulin on gene transcription through binding
to the IRE in the promoter sequences of target genes (26). Taken together, these data suggest
that insulin induces H3 modifications at the promoter and represses
the transcription of AQP9 gene in hepatocytes, which is mediated,
at least in part, via FOXO1-dependent alteration of chromatin
structure at the IRE locus. Further studies are required to clarify
to what extent insulin action is dependent on FOXO1-induced
epigenetic modifications. Additionally, the signaling pathways
involved in the regulation of FOXO1 by insulin requires further
elucidation.
In conclusion, the present study demonstrated that
insulin-induced transcriptional repression of AQP9 gene expression
in hepatocytes is associated with FOXO1-mediated H3 modifications
at the IRE locus of the AQP9 promoter. These findings
warrant further investigation of the clinical significance of
epigenetic regulation of AQP9 expression in treating metabolic
syndrome.
References
|
1
|
Verkman AS: Aquaporins. Curr Biol.
23:R52–R55. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Buffoli B: Aquaporin biology and nervous
system. Curr Neuropharmacol. 8:97–104. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Maeda N: Implications of aquaglyceroporins
7 and 9 in glycerol metabolism and metabolic syndrome. Mol Aspects
Med. 33:665–675. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Rojek AM, Skowronski MT, Füchtbauer EM, et
al: Defective glycerol metabolism in aquaporin 9 (AQP9) knockout
mice. Proc Natl Acad Sci USA. 104:3609–3614. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Jelen S, Wacker S, Aponte-Santamaría C, et
al: Aquaporin-9 protein is the primary route of hepatocyte glycerol
uptake for glycerol gluconeogenesis in mice. J Biol Chem.
286:44319–44325. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kuriyama H, Shimomura I, Kishida K, et al:
Coordinated regulation of fat-specific and liver-specific glycerol
channels, aquaporin adipose and aquaporin 9. Diabetes.
51:2915–2921. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Castro Parodi M, Farina M, Dietrich V,
Abán C, Szpilbarg N, Zotta E and Damiano AE: Evidence for
insulin-mediated control of AQP9 expression in human placenta.
Placenta. 32:1050–1056. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Rodríguez A, Catalán V, Gómez-Ambrosi J,
et al: Insulin- and leptin-mediated control of aquaglyceroporins in
human adipocytes and hepatocytes is mediated via the PI3K/Akt/mTOR
signaling cascade. J Clin Endocrinol Metab. 96:E586–E597. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Carbrey JM, Gorelick-Feldman DA, Kozono D,
Praetorius J, Nielsen S and Agre P: Aquaglyceroporin AQP9: solute
permeation and metabolic control of expression in liver. Proc Natl
Acad Sci USA. 100:2945–2950. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kamagate A, Qu S, Perdomo G, et al: FoxO1
mediates insulin-dependent regulation of hepatic VLDL production in
mice. J Clin Invest. 118:2347–2364. 2008.PubMed/NCBI
|
|
11
|
Matsumoto M, Pocai A, Rossetti L, Depinho
RA and Accili D: Impaired regulation of hepatic glucose production
in mice lacking the forkhead transcription factor Foxo1 in liver.
Cell Metab. 6:208–216. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Tsuchida A, Yamauchi T, Ito Y, et al:
Insulin/Foxo1 pathway regulates expression levels of adiponectin
receptors and adiponectin sensitivity. J Biol Chem.
279:30817–30822. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Xiao X, Mei ZC, Qiu LW, et al: Effect of
forkhead transcription factor 1 gene silencing on expression of
aquaporin 9 in normal human liver cells. Chin J Biologicals.
10:1157–1161. 2011.
|
|
14
|
Sharma S, Kelly TK and Jones PA:
Epigenetics in cancer. Carcinogenesis. 31:27–36. 2010. View Article : Google Scholar :
|
|
15
|
Gräff J, Kim D, Dobbin MM and Tsai LH:
Epigenetic regulation of gene expression in physiological and
pathological brain processes. Physiol Rev. 91:603–649. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kabra DG, Gupta J and Tikoo K: Insulin
induced alteration in post-translational modifications of histone
H3 under a hyperglycemic condition in L6 skeletal muscle myoblasts.
Biochim Biophys Acta. 1792:574–583. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gupta J and Tikoo K: Involvement of
insulin-induced reversible chromatin remodeling in altering the
expression of oxidative stress-responsive genes under hyperglycemia
in 3T3-L1 preadipocytes. Gene. 504:181–191. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hatta M and Cirillo LA: Chromatin opening
and stable perturbation of core histone: DNA contacts by FoxO1. J
Biol Chem. 282:35583–35593. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ornskov D, Nexo E and Sorensen BS: Insulin
induces a transcriptional activation of epiregulin, HB-EGF and
amphiregulin, by a PI3K-dependent mechanism: identification of a
specific insulin-responsive promoter element. Biochem Biophys Res
Commun. 354:885–891. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ge J, Zhai W, Cheng B, et al: Insulin
induces human acyl-coenzyme A: cholesterol acyltransferase1 gene
expression via MAP kinases and CCAAT/enhancer-binding protein α. J
Cell Biochem. 114:2188–2198. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yan C and Boyd DD: Histone H3 acetylation
and H3 K4 methylation define distinct chromatin regions permissive
for transgene expression. Mol Cell Biol. 26:6357–6371. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Fischle W, Wang YM and Allis CD: Histone
and chromatin cross-talk. Curr Opin Cell Biol. 15:172–183. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Cheung P, Tanner KG, Cheung WL,
Sassone-Corsi P, Denu JM and Allis CD: Synergistic coupling of
histone H3 phosphorylation and acetylation in response to epidermal
growth factor stimulation. Mol Cell. 5:905–915. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Matsuzaki H, Daitoku H, Hatta M, Tanaka K
and Fukamizu A: Insulin-induced phosphorylation of FKHR (Foxo1)
targets to proteasomal degradation. Proc Natl Acad Sci USA.
100:11285–11290. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Miao H, Zhang Y, Lu Z, Liu Q and Gan L:
FOXO1 involvement in insulin resistance-related pro-inflammatory
cytokine production in hepatocytes. Inflamm Res. 61:349–358. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Durham SK, Suwanichkul A, Scheimann AO,
Yee D, Jackson JG, Barr FG and Powell DR: FKHR binds the insulin
response element in the insulin-like growth factor binding
protein-1 promoter. Endocrinology. 140:3140–3146. 1999.PubMed/NCBI
|