Introduction
Basal-like breast cancers (BLBCs) are aggressive
malignancies that express the gene signatures of
basal/myoepithelial cells in mammary glands. In human breast
cancers, the basal-like subtype has been identified as a distinct
entity with a poor prognosis. Due to their underexpression of
estrogen receptor (ER), progesterone receptor (PR) and HER2, BLBCs
are unlikely to respond to the current targeted systemic therapy
(1–3).
To overcome the challenges in the treatment of
BLBCs, a novel avenue of investigation into the molecular basis for
this disease is urgently required. Recently, a number of studies
have demonstrated that long non-coding RNAs (lncRNAs) have pivotal
roles in the origination and progression of certain types of cancer
(4–6). While only a few studies on lncRNAs in
BLBCs have been reported, targeting lncRNAs with critical
regulating activities in BLBCs is a promising therapeutic strategy
for the future.
lncRNAs are RNA transcripts with no protein-coding
potential that are >200 bases in length, which have been
identified as high level regulators with multiple molecular
regulating mechanisms in gene networks (7–9). A
recent study revealed that numerous lncRNAs functionally contact
their adjacent mRNAs and take on the form of ‘lncRNA-mRNA pairs’ in
the regulatory network (10).
Through preliminary bioinformatics analysis
(http://genome.ucsc.edu/), the present study found
the novel lncRNA TCONS_00011636, which is located at 6p25 and is
transcribed from the upstream side of the FOXC1 promoter.
Therefore, it was named as FOXC1 promoter upstream transcript
(FOXCUT) by our group. FOXC1 is an important transcriptional factor
regulating a variety of biological processes, including
embryogenesis, tumorigenesis and epithelial-mesenchymal transition
(EMT) (11–13). Several recent studies have shown
that a high level of FOXC1 expression correlates with poor overall
survival in BLBCs, and that FOXC1 is associated with aggressive
phenotypes and increased cell proliferation and migration in breast
cancer cells (14–16). In other types of malignant tumors,
including pancreatic cancer, non-small cell lung cancer and
hepatocellular carcinoma (12–14,17,18),
overexpression of FOXC1 is strongly correlated with poor prognosis
of the patients. FOXC1 is now recognized as an important cancer
biomarker in BLBCs (15,16). However, the expression and function
of FOXCUT lncRNA in BLBCs and its association with the adjacent
mRNA FOXC1 remains to be determined.
The aim of the present study was to investigate the
expression profile of FOXCUT lncRNA in breast cancer tissues and
the functional role of FOXCUT in MDA-MB-231 and MDA-MB-468 human
BLBC cells in vitro.
Materials and methods
Patient samples
A total of 55 specimens were collected from 55
patients previously diagnosed with primary breast cancers at the
PLA General Hospital (Beijing, China) between 2007 and 2013.
Clinical classification was performed by immunohistochemical
studies for ER, PR, HER2, cytokeratin 5/6 and EGFR (19,20).
The series included examples from each of the molecular subtypes
based on their immunohistochemical surrogate: 16 luminal A-like
(ER+/PR+/HER2−), 8 luminal B-like (ER+/PR+/HER2+), 6 HER2-enriched
(ER-/PR-/HER2+) and 25 basal-like (ER-/PR-/HER2+/CK5/6+ or EGFR+).
The utilization of tumor material for research was approved by the
ethical committee of PLA General Hospital and written informed
consent was obtained from the patients or their families.
Cell line and cell culture
The human BLBC cell lines MDA-MB-231 and MDA-MB-468
were purchased from the American Type Culture Collection (Manassas,
VA, USA). The MDA-MB-231 cells were incubated in Dulbecco’s
modified Eagle’s medium (Hyclone, Logan, UT, USA) containing 10%
fetal bovine serum (FBS; Invitrogen, Carlsbad, CA, USA), and
MDA-MB-468 cells were cultured in RPMI-1640 medium (Hyclone)
supplemented with 10% FBS at 37°C with 5% CO2.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted from breast cancer tumor
tissues, matched adjacent normal tissues and breast cancer cells
using the TRIzol Total RNA reagent (Invitrogen Life Technologies,
Carlsbad, CA, USA). The primers were obtained from Sheng Gong
(Shanghai, China) and the sequences are presented in Table I. RT-qPCR was performed using the
SYBR PrimeScript RT-PCR kit (Takara, Ohtsu, Japan) in an Applied
Biosystems 7500 Fluorescent Quantitative PCR System (Applied
Biosystems, Foster City, CA, USA). The reaction mixtures were
incubated at 95°C for 30 sec, followed by 40 amplification cycles
of 95°C for 5 sec and 60°C for 34 sec. The comparative Ct method
was used to quantify relative expression of mRNA and lncRNA.
Expression levels of housekeeping gene β-actin were used to
normalize gene-of-interest expression. The expression levels of a
target gene in a patient were calculated as the ratio of the target
expression levels in tumor tissue to the target expression levels
in non-tumorous tissue (T/N).
 | Table IPrimers for real time polymerase chain
reaction analysis. |
Table I
Primers for real time polymerase chain
reaction analysis.
| Gene name | Forward | Reverse |
|---|
| β-actin |
5′-CCACTGGCATCGTGATGGA-3′ |
5′-CGCTCGGTGAGGATCTTCAT-3′ |
| FOXC1 |
5′-GGCGAGCAGAGCTACTACC-3′ |
5′-TGCGAGTACACGCTCATGG-3′ |
| FOXCUT |
5′-GTCGCACCGATGACTAACG-3′ |
5′-GCCCTGAAAGCCGAACTG-3′ |
Transfection of siRNA
The siRNA sequences were obtained from GenePharma
(Shanghai, China), including one negative control siRNA (NC siRNA)
sequence and two FOXCUT siRNA sequences. The target sequences are
presented in Table II. siRNA
transfection was performed with X-tremeGENE transfection reagent
(Roche, Mannheim, Germany). In brief, ~5% cells were plated in each
well of 12-well plates at least 24 h prior to transfection to
achieve 30–50% confluency. siRNA transfection was then performed
with X-tremeGENE transfection reagent (Roche) according to the
manufacturer’s instructions. Two days post-transfection, RNA
isolation, cell proliferation assay, scratch wound healing assay
and matrigel invasion assays were performed.
 | Table IISequences for small interfering RNA
analysis. |
Table II
Sequences for small interfering RNA
analysis.
| Gene name | Sense (5′-3′) | Antisense
(5′-3′) |
|---|
| FOXCUT si1 |
5′-GAAUGGAGAACUAAGACAAUUAUCT-3′ |
5′-AGAUAAUUGUCUUAGUUCUCCAUUCGG-3′ |
| FOXCUT si2 |
5′-CAGCCUCCCUCCUGUGUGUGCAGAG-3′ |
5′-CUCUGCACACACAGGAGGGAGGCUGCA-3′ |
Cell proliferation assay
Following transfection, cell proliferation was
assessed by a CellTiter 96® Aqueous
3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium
(MTS) assay kit (Promega) according to the manufacturer’s
instructions. MDA-MB-231 and MDA-MB-468 cells (2,000 cells per
well) in each group were plated in 96-well plates. MTS reagent (20
μl) was added to each well containing 100 μl culture medium. The
plate was incubated for 2 h at 37°C in a humidified, 5%
CO2 atmosphere. The plate was read at a wavelength of
490 nm using a SpectraMax M2 plate reader (Molecular Devices,
Sunnyvale, CA, USA).
Scratch wound healing assay
Prior to transfection, uniform wounds were scraped
into MDA-MB-231 and MDA-MB-468 cells grown on plastic six-well
plates using a pipette tip. The initial gap length (0 h) and the
residual gap length 48 h after wounding were calculated from
photomicrographs. Images were captured using a Olympus BX51 Clone
fluorescence microscope (Olympus Corp., Tokyo, Japan).
Matrigel invasion assays
A cell invasion assay was performed using modified
Boyden Chambers consisting of Transwell-precoated matrigel membrane
filter inserts with 8-mm pores in 24-well tissue culture plates (BD
Biosciences, Franklin Lakes, NJ, USA). Culturing medium containing
10% FBS in the lower chamber served as the chemoattractant. Cells
that had migrated through the filter were stained and counted. The
average migration rate was calculated as the increasing radius of
the entire cell population over time.
Statistical analysis
Differences between groups were analyzed using a
Student’s t-test. Correlation between gene expression levels were
studied using Pearson’s correlation. Statistical analyses were
performed using SPSS version 18.0 (International Business Machines,
Armonk, NY, USA). For all statistical analyses, P<0.05 was
considered to indicate a statistically significant difference.
Results
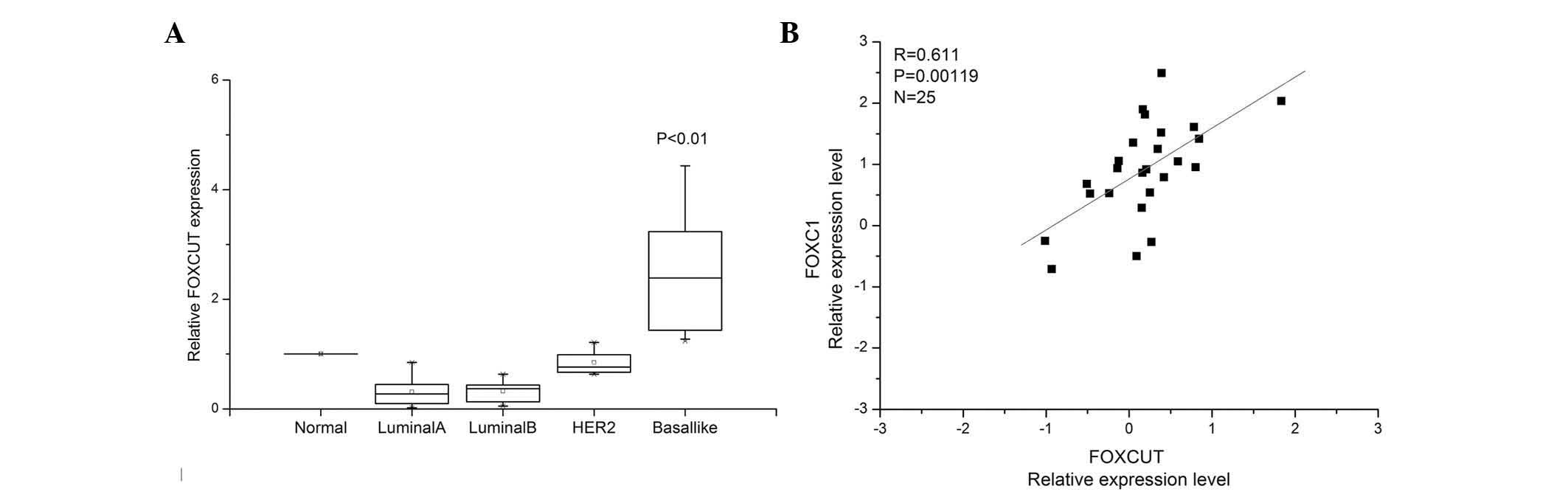
FOXCUT is overexpressed in BLBC tissue
specimens
The FOXCUT lncRNA expression levels were assessed in
a panel of paired specimens obtained from 55 patients with breast
cancer using RT-qPCR. The results revealed that the FOXCUT
expression levels in BLBC tumor tissues were significantly higher
than those in matched non-tumorous tissues. The expression levels
of FOXCUT were significantly higher in BLBCs than those in
non-basal like breast cancer subtypes (Fig. 1A, P<0.01). In addition, the
relative expression of FOXCUT was positively correlated with that
of FOXC1 in the BLBC tissue samples (Fig. 1B; R=0.611, P<0.01).
FOXC1 mRNA expression is suppressed by
FOXCUT siRNA in BLBC cell lines
In MDA-MB-231 and MDA-MB-468 cells, RNA interference
analysis was conducted to further clarify the correlation between
the expression of FOXCUT lncRNA and FOXC1 mRNA. RT-qPCR was
performed to evaluate the expression levels of FOXC1 mRNA and
FOXCUT lncRNA. The results showed that FOXCUT expression was
efficiently knocked down by FOXC1 siRNA (Fig. 2, P<0.05). In addition, FOXC1
expression was downregulated in the FOXCUT siRNA1 group compared
with that of the NC siRNA group (Fig.
2, P<0.05). This indicated that the expression of FOXC1 mRNA
may be modulated by FOXCUT lncRNA in BLBCs.
Knockdown of FOXCUT inhibits the cell
proliferation ability of MDA-MB-231 and MDA-MB-468 cells
To investigate the effects of FOXCUT knockdown on
the in vitro growth characteristics of the MDA-MB-231 and
MDA-MB-468 BLBC cell lines, an MTS assay was performed to assess
the cell proliferation ability. The results showed that cell growth
was inhibited in FOXCUT siRNA groups compared with that in the NC
siRNA group (Fig. 3,
P<0.05).
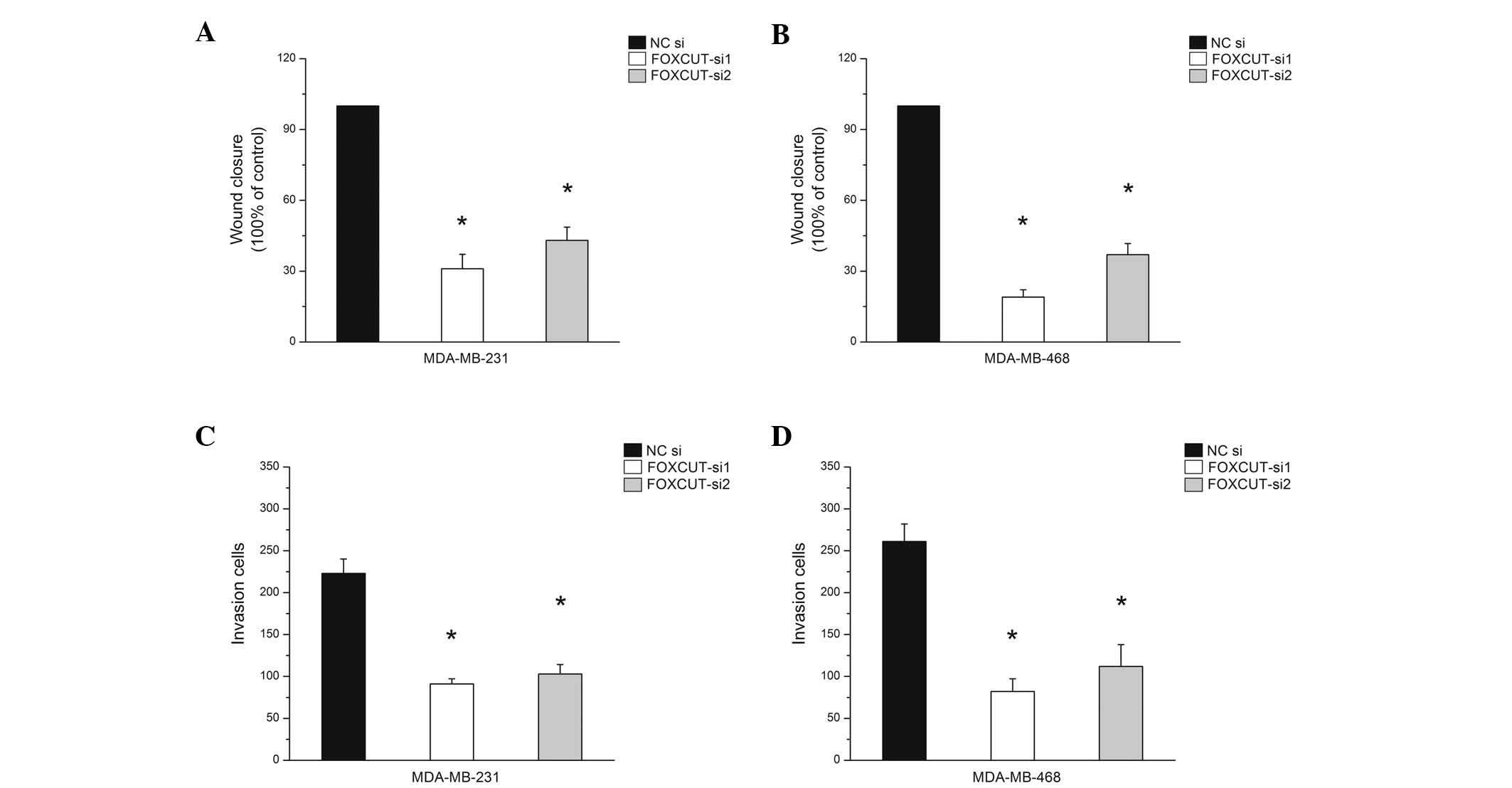
Knockdown of FOXCUT suppresses the
migration ability of MDA-MB-231 and MDA-MB-468 cells
To further identify the function of FOXCUT, a
scratch wound-healing assay and matrigel invasion assay were
performed following siRNA transfection. The results showed that the
migration capacity of the MDA-MB-231 and MDA-MB-468 cells was
markedly inhibited by FOXCUT siRNA (Fig. 4, P<0.05). The results of the
FOXCUT knockdown concurred with the effects of FOXC1 knockdown on
the in vitro growth characteristics of MDA-MB-231 and
MDA-MB-468 as shown above.
Discussion
Advances in high-throughput technologies have
resulted in the biological classification of breast cancer into
subtypes with distinct gene expression profiles, and BLBC is the
most aggressive subtype, with a unique gene-expression pattern of
basal/myoepithelial cells characteristics (1–3).
Previous studies regarding BLBC-associated genes
primarily focused on protein-coding genes. In recent years,
numerous studies have demonstrated the involvement of lncRNAs in
the development and progression of a number of types of malignant
tumors (4–6). In breast cancers, several lncRNAs
have been identified as novel biomarkers and therapeutic targets,
including HOTAIR, BC200 and CCAT2 (21–23).
However, the expression patterns and functional roles of
cancer-associated lncRNAs in BLBCs remain to be determined.
The present study reported a novel lncRNA, FOXCUT,
which may be a cancer-promoting gene responsible for the aggressive
phenotype in BLBCs. Through RT-qPCR investigation, it was
determined that the expression levels of lncRNA-FOXCUT were
remarkably elevated in BLBCs, yet not in other non-basal like
breast cancer subtypes. The expression levels of FOXCUT in BLBCs
were significantly higher than those in non-basal like breast
cancer subtypes, suggesting that lncRNA-FOXCUT may serve as a novel
specific biomarker in BLBCs.
Furthermore, In BLBC cell lines, it was revealed
that the knockdown of lncRNA-FOXCUT markedly inhibited cell
proliferation and migration. This indicated that FOXCUT is not only
a pure diagnostic marker in BLBCs, but also an important functional
regulator in the cell aggressiveness, similar to the well-known
lncRNA HOTAIR in breast cancer (21).
Cancer-associated lncRNAs may exert their regulating
activities through diverse mechanisms. Certain lncRNAs may perform
their functional roles by directly regulating their neighboring
protein coding genes, such as lncRNA PVT1 and protein coding gene
c-MYC (24,25). FOXC1 (FOXCUT) is an adjacent lncRNA
upstream of the FOXC1 promoter. These ncRNAs are called promoter
upstream transcripts (PROMPTs) and are often functionally
associated with the adjacent protein-coding transcripts (26–28).
Given that the function of the protein-coding gene FOXC1 and FOXCUT
lncRNA are involved in the progression of BLBCs by affecting the
cell proliferation and cell migration (14–16),
it is therefore speculated that FOXCUT lncRNA and FOXC1 mRNA may be
another functional lncRNA-mRNA pair that interact with each other
in BLBC. RT-qPCR results showed that the expression of FOXCUT
lncRNA was positively correlated with FOXC1 mRNA. Through RNA
interference analysis, it was determined that the knockdown of
lncRNA-FOXCUT clearly reduced the levels of FOXC1 mRNA expression,
which was in line with the inhibited cell growth rate and migration
ability. This indicated that FOXCUT lncRNA may promote the
aggressiveness of BLBC cells partly by regulating the expression of
protein-coding gene FOXC1. However, additional studies are required
for complete elucidation of the underlying mechanisms.
In conclusion, the present study was the first to
identify the expression and functional role of a novel lncRNA,
FOXCUT, and its association with the adjacent FOXC1 mRNA in BLBCs.
The results indicated that FOXCUT may be a potential diagnostic
marker and therapeutic target in the future.
References
|
1
|
Kreike B, van Kouwenhove M, Horlings H, et
al: Gene expression profiling and histopathological
characterization of triple-negative/basal-like breast carcinomas.
Breast Cancer Res. 9:R652007. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Rakha EA, Reis-Filho JS and Ellis IO:
Basal-like breast cancer: a critical review. J Clin Oncol.
26:2568–2581. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Carey LA, Perou CM, Livasy CA, et al:
Race, breast cancer subtypes, and survival in the Carolina Breast
Cancer Study. JAMA. 295:2492–2502. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Huarte M and Rinn JL: Large non-coding
RNAs: missing links in cancer? Hum Mol Genet. 19:R152–R161. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Spizzo R, Almeida MI, Colombatti A and
Calin GA: Long non-coding RNAs and cancer: a new frontier of
translational research? Oncogene. 31:4577–4587. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Tsai MC, Spitale RC and Chang HY: Long
intergenic noncoding RNAs: new links in cancer progression. Cancer
Res. 71:3–7. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Okazaki Y, Furuno M, Kasukawa T, et al:
Analysis of the mouse transcriptome based on functional annotation
of 60,770 full-length cDNAs. Nature. 420:563–573. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Guttman M, Amit I, Garber M, et al:
Chromatin signature reveals over a thousand highly conserved large
non-coding RNAs in mammals. Nature. 458:223–227. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Perez DS, Hoage TR, Pritchett JR, et al:
Long, abundantly expressed non-coding transcripts are altered in
cancer. Hum Mol Genet. 17:642–655. 2008. View Article : Google Scholar
|
|
10
|
Han L, Zhang K, Shi Z, et al: LncRNA
profile of glioblastoma reveals the potential role of lncRNAs in
contributing to glioblastoma pathogenesis. Int J Oncol.
40:2004–2012. 2012.PubMed/NCBI
|
|
11
|
Myatt SS and Lam EW: The emerging roles of
forkhead box (Fox) proteins in cancer. Nat Rev Cancer. 7:847–859.
2007. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Xia L, Huang W, Tian D, et al:
Overexpression of forkhead box C1 promotes tumor metastasis and
indicates poor prognosis in hepatocellular carcinoma. Hepatology.
57:610–624. 2013. View Article : Google Scholar
|
|
13
|
Xu ZY, Ding SM, Zhou L, et al: FOXC1
contributes to microvascular invasion in primary hepatocellular
carcinoma via regulating epithelial-mesenchymal transition. Int J
Biol Sci. 8:1130–1141. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sizemore ST and Keri RA: The forkhead box
transcription factor FOXC1 promotes breast cancer invasion by
inducing matrix metalloprotease 7 (MMP7) expression. J Biol Chem.
287:24631–24640. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ray PS, Wang J, Qu Y, et al: FOXC1 is a
potential prognostic biomarker with functional significance in
basal-like breast cancer. Cancer Res. 70:3870–3876. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ray PS, Bagaria SP, Wang J, et al:
Basal-like breast cancer defined by FOXC1 expression offers
superior prognostic value: a retrospective immunohistochemical
study. Ann Surg Oncol. 18:3839–3847. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang L, Gu F, Liu CY, Wang RJ, Li J and Xu
JY: High level of FOXC1 expression is associated with poor
prognosis in pancreatic ductal adenocarcinoma. Tumour Biol.
34:853–858. 2013. View Article : Google Scholar
|
|
18
|
Wei LX, Zhou RS, Xu HF, Wang JY and Yuan
MH: High expression of FOXC1 is associated with poor clinical
outcome in non-small cell lung cancer patients. Tumour Biol.
34:941–946. 2013. View Article : Google Scholar
|
|
19
|
Livasy CA, Karaca G, Nanda R, et al:
Phenotypic evaluation of the basal-like subtype of invasive breast
carcinoma. Mod Pathol. 19:264–271. 2006. View Article : Google Scholar
|
|
20
|
Nielsen TO, Hsu FD, Jensen K, et al:
Immunohistochemical and clinical characterization of the basal-like
subtype of invasive breast carcinoma. Clin Cancer Res.
10:5367–5374. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gupta RA, Shah N, Wang KC, et al: Long
non-coding RNA HOTAIR reprograms chromatin state to promote cancer
metastasis. Nature. 464:1071–1076. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Iacoangeli A, Lin Y, Morley EJ, et al:
BC200 RNA in invasive and preinvasive breast cancer.
Carcinogenesis. 25:2125–2133. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Redis RS, Sieuwerts AM, Look MP, et al:
CCAT2, a novel long non-coding RNA in breast cancer: expression
study and clinical correlations. Oncotarget. 4:1748–1762.
2013.PubMed/NCBI
|
|
24
|
Shtivelman E and Bishop JM: Effects of
translocations on transcription from PVT. Mol Cell Biol.
10:1835–1839. 1990.PubMed/NCBI
|
|
25
|
Guan Y, Kuo WL, Stilwell JL, et al:
Amplification of PVT1 contributes to the pathophysiology of ovarian
and breast cancer. Clin Cancer Res. 13:5745–5755. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Preker P, Nielsen J, Kammler S, et al: RNA
exosome depletion reveals transcription upstream of active human
promoters. Science. 322:1851–1854. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Jacquier A: The complex eukaryotic
transcriptome: unexpected pervasive transcription and novel small
RNAs. Nat Rev Genet. 10:833–844. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
28
|
Preker P, Almvig K, Christensen MS, et al:
PROMoter uPstream Transcripts share characteristics with mRNAs and
are produced upstream of all three major types of mammalian
promoters. Nucleic Acids Res. 39:7179–7193. 2011. View Article : Google Scholar : PubMed/NCBI
|