Introduction
The survival rate of patients with colorectal cancer
(CRC), which is one of the most common types of malignancy and the
third leading cause of cancer-associated mortality worldwide, is
delineated by a high rate of recurrence (1–3).
Mutations in certain tumor-suppressor genes and oncogenes have been
identified, including adenomatous polyposis coli, deleted in
colorectal cancer, mothers against decapentaplegic homolog 2
(Smad2), tumor protein 53 and kirsten rat sarcoma viral oncogene
homolog (4–6). These mutant genes have been used in
CRC therapy; however, their treatment effectivity is limited.
Therefore, further investigation of novel targeted therapeutics for
the treatment of CRC is required.
Previous studies have revealed that micro (mi)RNAs
can regulate tumor development by targeting their downstream genes
and have been identified as a novel mechanism which contributes to
the pathogenesis of CRC (7–9) has
been identified. These small RNAs, which are aberrantly expressed
in various types of cancer, act as oncogenes or tumor suppressors
and are thus candidate targets for cancer therapy.
In the present study, the results of five
microarray-based human colon cancer microRNA expression profiling
studies were examined (10–15),
comparing colon cancer tissue with normal tissue. The results
demonstrated that the miR-183/96/182 cluster was upregulated in the
colon cancer tissues. The miR-183/96/182 cluster contains three
members: miR-183, miR-96 and miR-182. It has been reported that
these miRNAs are located within a distance of 4 kb from each other
on the mouse chromosome 6qA3 and are transcribed in the same
direction. They are expressed coordinately and are important in the
sensorineural fates of cells in the mouse inner ear (10,11).
This miRNA cluster also has a significant role in the maintenance
and survival of hair cells and post-mitotic photoreceptors of the
retina (12–14). In order to examine the roles of
these miRNAs in the pathogenesis of CRC in the present study, a
microarray-based miRNA expression profiling study was performed to
compare miRNA expression levels in colon cancer and normal tissues.
Furthermore, the present study aimed to investigate the importance
of miR-183/182/96 in the proliferation of CRC cells using ASO-based
miRNA inhibitors.
Materials and methods
Cell culture
HT-29 and LoVo human colon cancer cell lines were
purchased from the American Type Culture Collection (Manassas, VA,
USA). The cells were maintained in Dulbecco’s modified Eagle’s
medium (DMEM; Life Technologies, Carlsbad, CA, USA) with 10% fetal
calf serum (FCS; Sigma-Aldrich), 100 U/ml penicillin and 100
μg/ml streptomycin (Life Technologies). All the cells were
maintained at 37°C under an atmosphere of 5% CO2 and 95%
air.
Patient samples
A total of eight paired human colon cancer tissue
and corresponding adjacent normal tissue samples were obtained from
randomly selected cancer patients at the Department of General
Surgery, The 15th Hospital of People’s Liberation Army (Xinjiang,
China) and all of the diagnoses were pathologically confirmed.
Written informed consent was obtained from each patient involved in
the present study prior to surgery and all procedures were reviewed
by the Joint Ethics Committee of the 15th Hospital of People’s
Liberation Army and performed in accordance with national
guidelines.
Literature search for studies that
examined the expression of miR-183/96/182 in colon cancer
tissues
The Pubmed database (http://www.ncbi.nlm.nih.gov/pubmed) was searched to
identify eligible studies that determine the association between
miR-183/96/182 and colon cancer. Search terms, including ‘miR-183’,
‘miR-96’, ‘miR-182’ and ‘colon cancer’ were used for the literature
search. The selected studies met the requirement that the
expression levels of miR-183/96/182 were quantitated in human colon
cancer and normal tissues. The clinical characteristics of these
studies were extracted and the fold changes of the expression
levels of the three miRNAs in colon cancer tissues were then
compared with those in normal tissue.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) analysis
Total RNAs were extracted from the tissues using
TRIzol reagent (Invitrogen Life Technologies, Carlsbad, CA, USA),
and miRNA were reverse transcribed using the miRCURY LNATM
Universal cDNA Synthesis kit II (Exiqon, Vedbak, Denmark). RT-qPCR
was then performed on an ABI Prism 7900 Sequence Detection system
(Applied Biosystems, Foster City, CA) in a 10 μl PCR
reaction mix, including 0.67 μl RT product, 1X SYBR Green
PCR master mix (Invitrogen) and 1 μl (25 ng) of both forward
and reverse primers. The reactions were incubated in triplicates in
a 96-well optical plate at 95°C for 10 min, followed by 40 cycles
of 95°C for 15 sec and 60°C for 1 min. U6 snRNA levels were used as
an endogenous control. The primers used for the RT-qPCR were
synthesized by Invitrogen Life Technologies as follows: Forward:
5′-GCGCCTATGGCACTGGTAGAA-3′ and reverse: 5′-TGCAGGGTCCGAGGTATTCG-3′
for miR-183; forward: 5′-TTTGGCACTAGCACAT-3′ and reverse:
5′-GAGCAGGCTGGAGAA-3′ for miR-96; forward:
5′-CGGCGGTTTGGCAATGGTAGAACT-3′ and reverse:
5′-CCAGTGCAGGGTCCGAGGTAT-3′ for miR-182; and forward:
5′-CGCTTCGGCAGCACATATACTA-3′ and reverse:
5′-CGCTTCACGAATTTGCGTGTCA-3′ for U6 snRNA. Data processing was
conducted using the SDS software (v2.1) (Applied Biosystems) and
the expression levels of the miRNAs were then calculated using the
2−ΔΔct method after normalized to the levels of
U6snRNA.
MTT assay
The synthesized RNA duplexes of antiscram-bled
(ASO-miR-NC), ASO-miR-183, ASO-miR-182 and ASO-miR-96 mimics were
obtained from GeneChem (Shanghai, China). Following transient
transfection of miRNA inhibitors, the HT-29 or LoVo cells were
seeded into 96-well plates at 1,500 cells/well and MTT
(Sigma-Aldrich, St. Louis, MO, USA) assays were performed daily for
72 h. In this assay, the medium was replaced with fresh medium
containing 0.5 mg/ml MTT for 4 h and then carefully removed.
Subsequently, 150 μl dimethyl sulfoxide (Sigma-Aldrich) was
added to each well and mixed for 10 min, and the optical density at
490 nm was determined using an enzyme linked immunosorbent assay
reader (BioTek Instruments, Winooski, VT, USA). With the MTT we
designed eight groups (A-H), which contained ASO-miR-183,
ASO-miR-96, ASO-miR-182, either alone or in combinations of two or
all three, as well as a ASO-miR-NC control group.
Colony formation assay
The cells were seeded into a 12-well plate at a
density of 200 cells/well following transfection. The medium was
changed every 3 days. After ~10 days, the majority of the cell
clones contained >50 cells. The colonies were then washed with
1X phosphate-buffered saline and stained with crystal violet
(Fisher Scientific, Pittsburgh, PA, USA) for ~5 min. Finally,
images of the colonies were captured using a Nikon Eclipse E800
microscope (Nikon, Tokyo, Japan) and the number of colonies was
counted. The colony formation rate (%) = (number of clones) /
(number of seeded cells) × 100.
Western blot analysis
Western blot analysis was performed, as previously
described (15). Rabbit polyclonal
anti-Ki-67 (ab15580) and Rabbit polyclonal anti p-protein kinase B
(Akt; ab66138) were obtained from Abcam (Cambridge, MA, USA). Mouse
monoclonal antibodies against Bcl 2-associated X protein (Bax;
sc-20067) and p53 (sc-126) and horseradish peroxidase conjugated
goat anti-mouse IgG (sc-2005) and goat anti-rabbit IgG (sc-2004)
were purchased from Santa Cruz Biotechnology, Inc. (Dallas, TX,
USA). The membranes were incubated with primary antibodies at 4 ºC
overnight, followed by incubation with horseradish
peroxidase-conjugated secondary antibodies. Proteins were then
detected using an enhanced chemiluminescence kit (GE Healthcare
Life Sciences, Piscataway, NJ, USA). LabWork 4.0 software was used
to measure the band intensities of the blots.
Statistical analysis
All data are expressed as the mean ± standard
deviation. The difference between groups was determined using
two-tailed Student’s t-test. Statistical analyses were performed
using Micrsoft Excel 2013 (Microsoft Corp., Redmond, WA, USA).
P<0.05 was considered to indicate a statistically significant
difference.
Results
All miR-183/96/182 cluster members are
upregulated in colon cancer
The miR-183/96/182 cluster was located on the region
of human chromosome 7q and the miRNAs were transcribed in the same
direction. It has been suggested that this cluster is unregulated
in colon cancer tissues. To confirm the expression levels of the
members of this gene cluster in colon cancer, a total of five
previous studies (Table I)
(16–20), which investigated miRNA expression
in colon cancer, were examined and the fold changes of these three
miRNAs in colon cancer tissues were compared with those in normal
tissue. The clinical characteristics of these studies were
extracted and are listed in Table
I. All three miRNAs had ~2-fold changes in the colon cancer
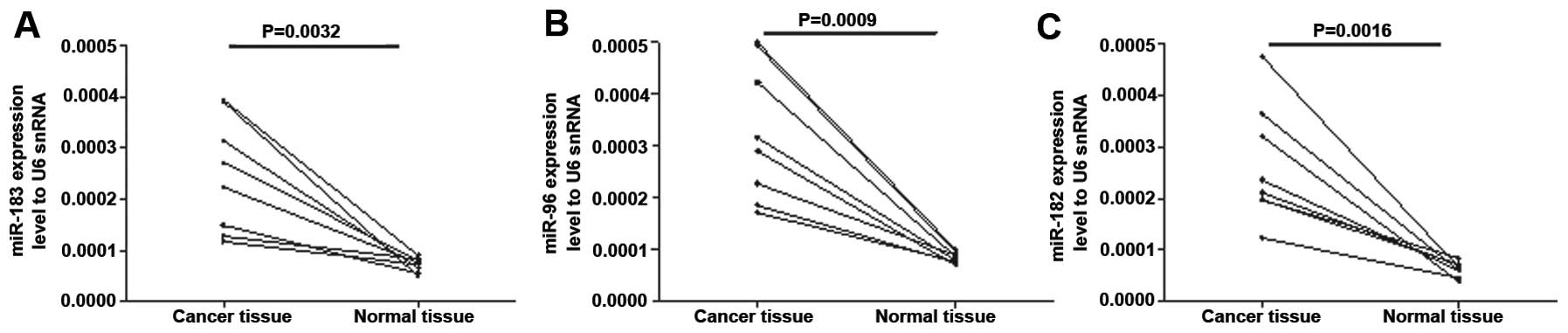
tissues according to the five microarray results. To confirm these
findings, RNAs were extracted from eight colon cancer samples with
paired adjacent normal colon tissues and these were analyzed by
RT-qPCR (Fig. 1A–C). Consistent
with the Table I data, the results
demonstrated that miR-183, miR-96 and miR-182 were expressed at
relatively high levels in the colon cancer tissues.
 | Table IFive microarray-based human colon
cancer differential miRNA expression profiling studies (colon
cancer tissue, vs. normal tissue). |
Table I
Five microarray-based human colon
cancer differential miRNA expression profiling studies (colon
cancer tissue, vs. normal tissue).
| Study (Ref) | Year | Origin | Period | Cancer type | No. samples
(cancer/normal) | Platform | Total | Upregulated | Downregulated | miR-183/96/182
cluster |
|---|
| (16) | 2009 | USA | Beginning in
1995 | Colon cancer | 108 (80/28) | Illumina
miRNA
Detection Platform | 39 | 17 | 22 |
miR-182↑2.21
miR-183↑2.59
miR-96 ↑2.04 |
| (17) | 2012 | Norway | No report | Colorectal
cancer | 8 (8/8) Paired | Ilumina Sequencing
Technology | 37 | 18 | 19 | miR-96 ↑3.2 |
| (18) | 2012 | Italy | No report | Colorectal
cancer | 19 (19/19)
Paired | Gene Chip miRNA Array
(www.affymetrix.com) | 42 | 25 | 17 |
miR-182↑3.694
miR-183↑3.064 |
| (19) | 2009 | USA | No report | Metastatic colorectal
cancer | 49 (45/4) | mirVana Bioarray
(Ambion, Version1) | 37 | 22 | 15 | miR-182↑2.8
miR-183↑1.8
miR-96 ↑2.0 |
| (20) | 2006 | Spain | No report | Colorectal
cancer | 12 (12/12) Paired 15
cancer cell lines | BRB-Array (Colorectal
cancer tissue and cell lines) | 13 | 4 | 9 |
miR-182↑3.41
miR-183↑1.74
miR-96 ↑1.99 |
Simultaneous knockdown of the expression
of the miR-183/96/182 cluster efficiently inhibits HT-29 and LoVo
cell viability compared with inhibiting the miRs alone
It has been demonstrated that miR-183, 96 and 182
act as tumor oncogenes based on their high expression levels in
colon cancer tissues (21).
Therefore, the present study used ASO-miRs to knockdown the
expression of the miR-183/96/182 cluster. It has also been
suggested that miR-183, miR-96 and miR-182 have similar sequences
and are highly conserved across species, therefore, raising the
question of whether the three miRNAs acted coordinately or
competitively to regulate the growth phenotype of colon cancer. To
address this question, the present study designed eight groups,
defined as groups A-H, of ASO-miRNAs containing different
concentrations of ASO-miR-183, ASO-miR-96, ASO-miR-182 and
ASO-miR-NC, either alone or in combination (Fig. 2). Subsequently, these ASO mimics
were transfected into the HT-29 cells and the cell viability was
measured using an MTT assay. As Fig.
2B shows, the HT-29 cell viability was classified into four
levels. Arbitrary knockdown of two members of the miR-183/96/182
cluster (level 3) caused a reduction in HT-29 cell viability
compared with the arbitrary knockdown of each alone (level 2).
Furthermore, knockdown of all three members of the miR-183/96/182
cluster (group H) efficiently inhibited HT-29 cell viability (level
4) compared with the others (groups A-G). The same results were
observed in the LoVo cells (Fig.
2C). These results suggested that simultaneous knockdown of the
expression of the miR-183/96/182 cluster efficiently inhibited
colon cancer cell viability.
Simultaneous knockdown of the
miR-183/96/182 cluster expression efficiently inhibits HT-29 cell
colony formation ability compared with inhibition of the miRs
alone
In the present study, an MTT assay was used to
detect the colon cancer cell viability 72 h (Fig. 2B and C) after transfection with the
ASO-miRNAs. To further confirm that simultaneous knockdown of all
the members of the miR-183/96/182 cluster efficiently inhibited
colon cancer cell viability compared with knockdown of the miRs
alone, a colony formation assay was performed. According to the
design of the MTT assay, eight transfection groups were used. As
shown in Fig. 3A, the colony
formation rates of the HT-29 cells transfected with ASO-miR-183,
ASO-miR-96 or ASO-miR-182 (groups A, B, C and D) were lower
compared with ASO-miR-NC (A). In addition, the cells simultaneously
transfected with ASO-miR-183 and ASO-miR-96, ASO-miR-183 and
ASO-miR-182 or ASO-miR-182 and ASO-miR-96 (groups E, F and G) had a
higher colony formation rate compared with those simultaneously
transfected with miR-183, miR-96 and miR-182 (group H). These
results were consistent with those of the MTT assay, which
demonstrated that simultaneous knockdown of the expression of the
miR-183/96/182 cluster efficiently reduced HT-29 cell proliferation
compared with inhibition of the miRs alone.
Simultaneous knockdown of the expression
of the miR-183/96/182 cluster efficiently regulates key
proliferation of colon cancer cells and expression of the apoptotic
protein marker
To investigate the effect of simultaneous knockdown
of the expression of the miR-183/96/182 cluster on the
proliferation/apoptotic signaling pathway, Ki-67, phosphorylated
(p)-Akt, Bax and TP53 were examined by western blot analysis. In
the HT-29 and LoVo cells, the combined inhibition of the miR-183
cluster increased the activated expression of wild-type p53 and Bax
and decreased the expression of p-Akt and the cell proliferation
marker Ki-67 (Fig. 4).
Collectively, the observation of induced apoptosis and decreased
proliferation resulting from pooled knockdown of the miR-183
cluster in colon cells implied that treatment of colon cancer
tumorigenesis using miRNA as a target in an miRNA-cluster-dependent
manner may efficiently reduce colon cancer cell proliferation.
Discussion
miRNAs, ~22 nt in length, are a novel class of
regulatory molecules with the ability to control the expression
levels of thousands of genes and appear to decrease the expression
of proteins by increasing the degradation or suppressing the
translation of mRNA (22).
Accumulating evidence indicates that miRNAs also function as
oncogenes or tumor suppressor genes, which contribute to the
tumorigenesis of several types of cancer, including colon cancer
(23,24). In the present study, five eligible
studies containing 196 samples and 15 CRC cell lines were examined.
As listed in Table I, several
dysregulated miRNAs were found, and subsequent investigation
focused on the miR-183/96/182 cluster. Based on the data in
Table I, the average expression
levels of miR-183, miR-96 and miR-182 increased 2.30-, 2.31- and
3.03-fold, respectively, in colon cancer tissues compared with
normal tissue.
The human miR-183/96/182 cluster is located on human
chromosome 7q32.2. The combined expression of these miRNAs may
function in physiology and pathology, including tumor pathology.
The human miR-183/96/182 cluster has been demonstrated as being
overexpressed in several types of tumor and acting as an oncogene.
Han et al (25) suggested
that its overexpression is a marker for bladder cancer. Mihelich
et al (26) identified the
members of this cluster as having diagnostic and prognostic
implications in prostate cancer. In addition, Yamada et al
(27) identified two members of
the cluster, miR-96 and miR-183 serve as potential tumor markers of
urothelial carcinoma and Weeraratne et al (28) reported that the effects of the
miR-183/96/182 cluster converge to regulate cell survival,
proliferation and migration in medulloblastoma. The miR-183/96/182
cluster was also found to regulate oxidative apoptosis and
sensitize cells to chemotherapy in gliomas (16,25–29).
However, the effects of their coordinate expression on the
mechanisms of tumorigenesis and particularly the proliferation of
colon cancer remain to be fully elucidated. In the present study,
this cluster was overexpressed in colon cancer, which was in
accordance with a previous study (21). A series of transfection
oligo-nucleotides were designed to detect the effects of the
miR-183/96/182 cluster in colon cancer. Notably, these miRNAs were
observed to coordinately regulate the proliferation of colon cancer
cells with a synergistic effect, which was termed 1 × ASO-miRNA = 1
× cell proliferation inhibition in the present study, however 3 ×
ASO-miRNA >3 × cell proliferation inhibition (Fig. 5).
The results of the present study indicated that the
combined biological effects of the three miRNAs in the
miR-182/96/183 cluster possessed increased inhibitory properties
compared with each individual miRNA alone. They exhibited the same
directional transcription and highly conserved ‘seed sequences’ and
acted as a unit that significantly regulated the phonotype of the
colon cancer cells, similar to the results observed by Tang et
al in glioma (29). These
findings provided support that miRNAs, which reside in clusters in
the genome, function synergistically in cancer tumorigenesis. To
further explain these mechanisms, the present study used the
miRBASE database (http://www.mirbase.org/) to identify the targets of
miR-182/96/183. As shown in Table
II, 105 validated targets of miR-96, 81 targets of miR-182 and
90 targets of miR-183 were identified. However, only 20 targets
were simultaneously targeted by all three miRNAs, which may explain
why their simultaneous inhibition led to a synergistic increase in
cell proliferation inhibition compared with inhibition of the
miRNAs alone.
 | Table IIValidated targets of miR-182/96/183
in the miRBASE database. |
Table II
Validated targets of miR-182/96/183
in the miRBASE database.
Validated targets
|
|---|
miR-96
| miR-182
| miR-183
| miR-182/96/183
|
|---|
| HIF1A | COX8A | VAMP8 | JAK2 | FOXO3 | TP53 | GFI1 | PRPH2 | TECTA | ARRDC3 |
| PAK3 | EIF2C1 | BNIP3L | IL17A | NFIB | EGR1 | HMOX1 | NFIB | BTRC | FOXO1 |
| ARRDC3 | TBK1 | MYD88 | FOXO1 | PNLIP | EVI1 | FSCN1 | KLF4 | RET | ATOH1 |
| ARIH2 | CISH | PSAT1 | HMGA2 | FBXW7 | ROS1 | RDX | GJB6 | IRS1 | DICER1 |
| RNASEH2A | USF2 | MTSS1 | SAG | PRMT5 | COX8A | MSH6 | PIK3CA | EGR1 | NEUROD4 |
| HTR1B | FOXO3 | HMG2L1 | BCL2L11 | RDX | ZEB2 | NEUROD4 | EIF2C1 | SOX2 | AKT1 |
| RGS2 | FXR1 | POMC | MYC | RAC1 | ERBB2 | GEMIN4 | MYB | TWIST1 | PTEN |
| FOXO1 | GFI1 | RHOD | BCL2 | MITF | PAK1 | MYO15A | AKAP12 | MLH1 | CASP2 |
| STAT6 | LTBP1 | SLC12A5 | AKT1 | DOK4 | RNASEN | EGFR | KITLG | C2 | ZEB1 |
| MYCN | GEMIN4 | BRAP | EGFR | ADCY6 | FRAP1 | IFI44 | DOK4 | HBEGF | TP53 |
| ATOH1 | SLC1A1 | IL23A | ARRDC3 | CDKN1A | SLC1A1 | ATOH1 | BBS9 | ERBB2 | EIF2C2 |
| HPRT1 | FBN1 | MYC | MYB | NAMPT | SLC7A5 | PI3 | TMC1 | RASA1 | COX8A |
| DICER1 | MYBL1 | PITPNM1 | ENPP3 | E2F3 | CCND1 | TIA1 | BRAF | ZEB1 | EIF2C1 |
| NEUROD4 | GEMIN5 | NCALD | DNTT | CDC42 | ZEB1 | MOS | PRB1 | MSH2 | CCND1 |
| SSSCA1 | RIT2 | ADCY6 | MTSS1 | PAK3 | RGS17 | APC | CTNNB1 | CCL27 | MOS |
| AKT1 | RNASEN | KRAS | KIT | SIRT1 | EIF2C3 | CASP2 | CCND1 | JAK2 | SAG |
| HNF1A | IRS1 | TIA1 | SSSCA1 | BHLHB5 | ZNF828 | RNPC3 | ITGB1 | EZR | FBXW7 |
| HOMER1 | DKK2 | CREBZF | MYCN | BDNF | IRAK2 | SLC26A4 | DDX20 | EIF2C2 | DOK4 |
| CAPNS1 | STAT3 | IL8 | MOCOS | NRIP1 | BAX | TNF | COL11A2 | ELSPBP1 | ADCY6 |
| SOCS2 | BCR | SIRT1 | DICER1 | HBEGF | CCL27 | PDCD4 | TP53 | TIAM1 | MITF |
| PTEN | SCPEP1 | FABP4 | EP300 | IL17F | IL2 | POU4F3 | FBXW7 | DFNB59 | |
| TGFB1 | ZEB2 | PRMT5 | NEUROD4 | RARG | EIF2C2 | DICER1 | HMGA2 | MITF | |
| KLK3 | FOSB | CCND1 | PRB1 | BRCA1 | CASP2 | SFRS2 | PTEN | STMN1 | |
| CASP2 | EIF2C2 | MOS | PCNA | MOS | ATOH1 | SAG | COX8A | ATP8A2 | |
| FRAP1 | TBP | PTPRR | CREB1 | RELA | TNF | BIRC5 | NRIP1 | KRAS | |
| ALK | SAG | DOK4 | CD38 | SOX6 | CTTN | IDH2 | KIF2A | ARRDC3 | |
| PRPH2 | BBC3 | SIP1 | PTEN | SLC12A2 | EIF2C1 | ADCY6 | GJB2 | AMACR | |
| CDKN1A | MAPK8 | FSCN1 | | | | IGF2 bp1 | RHOD | NPC1 | |
| FMR1 | MCL1 | FBXW7 | | | | NTRK1 | AKT1 | FOXO1 | |
| BCL2L11 | CDKN1B | DDIT4 | | | | GRB2 | FBN1 | CA1 | |
| ZEB1 | TP53 | SOX9 | | | | | | | |
| GADD45A | EPB41L3 | E2F3 | | | | | | | |
| DDX20 | CDH17 | CREB1 | | | | | | | |
| PPIA | CACNA2D2 | MITF | | | | | | | |
| MAP4K1 | NR3C1 | GPHN | | | | | | | |
In conclusion, the present study demonstrated that
increased expression of the miR-183/96/182 cluster was implicated
in human colon cancer. Knockdown of the miR-183/96/182 cluster
inhibited the survival of colon cancer cells and knockdown of the
miR-183/96/182 cluster enhanced the anticancer proliferation effect
more efficiently than knockdown of each alone. The co-expression of
miRNA cluster ASOs may be a pleiotropic target for colon cancer
therapy.
References
|
1
|
Weir HK, Thun MJ, Hankey BF, et al: Annual
report to the nation on the status of cancer, 1975–2000, featuring
the uses of surveillance data for cancer prevention and control. J
Natl Cancer Inst. 95:1276–1299. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Greenlee RT, Murray T, Bolden S and Wingo
PA: Cancer statistics, 2000. CA Cancer J Clin. 50:7–33. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Jemal A, Bray F, Center MM, et al: Global
cancer statistics. CA Cancer J Clin. 61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Fearon ER and Vogelstein B: A genetic
model for colorectal tumorigenesis. Cell. 61:759–767. 1990.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Cho KR and Vogelstein B: Genetic
alterations in the adenoma-carcinoma sequence. Cancer.
70:1727–1731. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Vogelstein B and Kinzler KW: The multistep
nature of cancer. Trends Genet. 9:138–141. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bhatti I, Lee A, Lund J and Larvin M:
Small RNA: a large contributor to carcinogenesis? J Gastrointest
Surg. 13:1379–1388. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Schetter AJ and Harris CC: Alterations of
microRNAs contribute to colon carcinogenesis. Semin Oncol.
38:734–742. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wu WK, Law PT, Lee CW, et al: MicroRNA in
colorectal cancer: from benchtop to bedside. Carcinogenesis.
32:247–253. 2011. View Article : Google Scholar
|
|
10
|
Li H, Kloosterman W and Fekete DM:
MicroRNA-183 family members regulate sensorineural fates in the
inner ear. J Neurosci. 30:3254–3263. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhu Q, Sun W, Okano K, et al: Sponge
transgenic mouse model reveals important roles for the microRNA-183
(miR-183)/96/182 cluster in postmitotic photoreceptors of the
retina. J Biol Chem. 286:31749–31760. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Xu S, Witmer PD, Lumayag S, Kovacs B and
Valle D: MicroRNA (miRNA) transcriptome of mouse retina and
identification of a sensory organ-specific miRNA cluster. J Biol
Chem. 282:25053–25066. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Krol J, Busskamp V, Markiewicz I, et al:
Characterizing light-regulated retinal microRNAs reveals rapid
turnover as a common property of neuronal microRNAs. Cell.
141:618–631. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Weston MD, Pierce ML, Jensen-Smith HC, et
al: MicroRNA-183 family expression in hair cell development and
requirement of microRNAs for hair cell maintenance and survival.
Dev Dyn. 240:808–819. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wu M, Huang C, Gan K, et al: LRRC4, a
putative tumor suppressor gene, requires a functional leucine-rich
repeat cassette domain to inhibit proliferation of glioma cells in
vitro by modulating the extracellular signal-regulated
kinase/protein kinase B/nuclear factor-kappaB pathway. Mol Biol
Cell. 17:3534–3542. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sarver AL, French AJ, Borralho PM, et al:
Human colon cancer profiles show differential microRNA expression
depending on mismatch repair status and are characteristic of
undifferentiated proliferative states. BMC Cancer. 9:4012009.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hamfjord J, Stangeland AM, Hughes T, et
al: Differential expression of miRNAs in colorectal cancer:
comparison of paired tumor tissue and adjacent normal mucosa using
high-throughput sequencing. PLoS One. 7:e341502012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Piepoli A1, Tavano F, Copetti M, et al:
Mirna expression profiles identify drivers in colorectal and
pancreatic cancers. PLoS One. 7:e336632012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Arndt GM1, Dossey L, Cullen LM, et al:
Characterization of global microRNA expression reveals oncogenic
potential of miR-145 in metastatic colorectal cancer. BMC Cancer.
9:3742009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Bandrés E1, Cubedo E, Agirre X, et al:
Identification by Real-time PCR of 13 mature microRNAs
differentially expressed in colorectal cancer and non-tumoral
tissues. Mol Cancer. 5:292006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Sarver AL, Li L and Subramanian S:
MicroRNA miR-183 functions as an oncogene by targeting the
transcription factor EGR1 and promoting tumor cell migration.
Cancer Res. 70:9570–9580. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bartel DP: MicroRNAs: genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Gregersen LH, Jacobsen AB, Frankel LB, et
al: MicroRNA-145 targets YES and STAT1 in colon cancer cells. PLoS
One. 5:e88362010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Tazawa H, Tsuchiya N, Izumiya M and
Nakagama H: Tumor-suppressive miR-34a induces senescence-like
growth arrest through modulation of the E2F pathway in human colon
cancer cells. Proc Natl Acad Sci USA. 104:15472–15477. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Han Y, Chen J, Zhao X, et al: MicroRNA
expression signatures of bladder cancer revealed by deep
sequencing. PLoS One. 6:e182862011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Mihelich BL, Khramtsova EA, Arva N, et al:
miR-183-96-182 cluster is overexpressed in prostate tissue and
regulates zinc homeostasis in prostate cells. J Biol Chem.
286:44503–44511. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yamada Y, Enokida H, Kojima S, et al:
MiR-96 and miR-183 detection in urine serve as potential tumor
markers of urothelial carcinoma: correlation with stage and grade,
and comparison with urinary cytology. Cancer Sci. 102:522–529.
2011. View Article : Google Scholar
|
|
28
|
Weeraratne SD, Amani V, Teider N, et al:
Pleiotropic effects of miR-183~96~182 converge to regulate cell
survival, proliferation and migration in medulloblastoma. Acta
Neuropathol. 123:539–552. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Tang H, Bian Y, Tu C, et al: The
miR-183/96/182 cluster regulates oxidative apoptosis and sensitizes
cells to chemotherapy in gliomas. Curr Cancer Drug Targets.
13:221–231. 2013. View Article : Google Scholar
|