Introduction
Chronic hepatitis C infection (CHC), which is
present worldwide, has been identified to increasingly contribute
to health care expenditure, morbidity and mortality (1). Although a strong correlation has been
observed between stage and prognosis in CHC, current CHC screening
methods have significant limitations (2). Although hepatitis C virus (HCV)-RNA
is currently the ‘gold standard’ for the diagnosis of HCV infection
and is frequently used for assessing the efficacy of anti-viral
agents, HCV viral load does not necessarily accurately correlate
with the severity and progression of the disease (3). Thus, the investigation of novel
sensitive and specific biomarkers for the early diagnosis of CHC is
required.
MicroRNAs (miRNAs) are single-stranded RNAs of
endogenous origin with a length of ~22 nucleotides, which function
in the post-transcriptional regulation of gene expression (4). This regulation is effected via the
mediation of mRNA degradation and/or translational blockade
(5); thus, they possess important
roles in a variety of physiological and pathological processes
(6,7). Of note, miRNAs have been identified
as crucial in the pathogenesis of HCV infection-associated liver
disease; dysregulations of miRNA have been demonstrated to be
involved in the modulation of HCV replication (8–10),
translation (11), gene expression
(12,13) and in the control of its response to
interferon (IFN) (14).
With their stability in circulation, relative
convenience of extraction, quantification and detection and the
power of polymerase chain reaction (PCR), circulating miRNAs can be
used effectively as noninvasive biomarkers (15). miRNAs are used in a wide range of
human diseases, including tumors, cardiac injury, tissue injury,
sepsis and pregnancy and offer potential for earlier diagnosis,
disease progression monitoring and improved precision of
personalized medication (15). A
previous study observed complementation between miR-196a and the
nonstructural (NS) 5A coding region of the HCV JFH1 genome; in
addition, IFN-β treatment led to significant miR-196 induction in
the Huh-7 human hepatoma cell line and in primary murine
hepatocytes (12). This suggested
a significant role for miR-196a in the modulation of HCV expression
and the therapeutic response of antiviral agents in human
hepatocytes. A previous study identified that miR-196a inhibited
HCV expression in the HCV replicon cell line and J6/JFH1 HCV cell
culture system, in addition to targeting the HCV genome and the
3′-untranslated region of Bach1 mRNA (13). The latter leads to upregulation of
the heme oxygenase (decycling) 1 gene, a key cytoprotective enzyme
that generates antioxidative and anti-inflammatory molecules
(13). Thus, miR-196a may
represent an important factor in the pathogenesis of HCV infection.
It was suggested that upregulation of miR-196a may be used in a
novel strategy to prevent or treat HCV infection, and miR-196a may
be valuable in the diagnosis and management of this disease
(13). However, the clinical
implications of aberrant miR-196a expression and the value of
circulating miR-196a in the diagnosis and management of chronic HCV
infection require further investigation.
Using an in vitro cell culture model and
serum samples from clinical patients, the present study aimed to
investigate the use of miR-196a as a novel candidate serum
biomarker for early CHC diagnosis.
Materials and methods
Cell culture
HepG2 cells, purchased from the American Type
Culture Collection (Manassas, VA, USA), were cultured in minimum
essential medium (GE Healthcare Life Sciences, Logan, UT, USA)
supplemented with 10% (v/v) fetal calf serum, 2 mmol/l glutamine,
100 U/ml penicillin and 100 μg/ml streptomycin (all from Gibco Life
Technologies, Carlsbad, CA, USA) at 37°C in a humidified
chamber.
Construction of the Ad-HCV core
adenovirus and the infection of HepG2 cells
Using the Stratagene AdEasy system (Agilent
Technologies, Inc., La Jolla, CA, USA), the Ad-HCV core adenovirus
and the control Ad-green fluorescent protein adenovirus were
constructed as previously reported (16). The infection of HepG2 cells (at a
multiplicity of infection of 50) and the evaluation of the
infection efficiency were performed according to the same study
(16). Cells were then harvested
for miRNA array, total RNA, protein analysis and
immunohistochemistry.
miRNA microarray analysis
miRNA microarray analysis was performed as
previously described (17).
Briefly, following the extraction of total RNA from the HepG2-HCV
and HepG2-control cells using TRIzol (Invitrogen Life Technolodies,
Carlsbad, CA), miRNA arrays (Affymetrix, Inc., Santa Clara, CA,
USA) were labeled and hybridized according to the manufacturer’s
instructions. The comparisons of miRNA expression data between
groups were performed with ComparativeMarkerSelection suite in
GenePattern software, version 10 (http://www.broadinstitute.org/cancer/software/genepattern).
Western blot analysis
Proteins extracted by the M-PER Mammalian Protein
Extraction Reagent (Cell Signaling Technology, Inc., Danvers, MA,
USA) were resolved on 10% SDS-PAGE gels (Bio-Rad Laboratories,
Inc., Hercules, CA, USA) and transferred to polyvinylidene fluoride
membranes (Pierce Biotechnology, Inc., Rockford, IL, USA). The
monoclonal mouse anti-Flag (the Ad-HCV core was tagged with 3X
Flag) primary antibody (1:500; ab49763; Abcam, Cambridge, UK) was
used overnight at 4°C and the horseradish peroxidase-linked rabbit
anti-mouse IgG (1:10,000; ab97046; Abcam) was used at room
temperature for 1 h as the secondary antibody. The monoclonal mouse
GAPDH antibody (1:1,000; ab8245; Abcam) was used overnight at 4°C
as a loading control. Blots were developed using Supersignal
WestPico chemiluminescent substrate (Pierce Biotechnology, Inc.),
imaged and analyzed using the Bio-Rad ChemiDoc XRS Gel Imaging
System (Bio-Rad Laboratories, Inc.).
Ethics statement
The experiments involving human participation were
conducted in accordance with the Declaration of Helsinki of 1975
and were approved by the Medical Ethics Committee on human research
of the First Affiliated Hospital of Chongqing Medical University
(Chongqing, China). All participants provided written informed
consent prior to enrollment.
Serum collection and storage
Blood samples from the patients in the emergency
department were collected, and through a two-step centrifugation
(10 min of 820 × g, then 10 min of 16,000 × g at 4°C), the
supernatant was transferred to RNase/DNase-free tubes and stored at
‒80°C within 1 h of collection.
Serum chemistry
Serum levels of alanine aminotransferase (ALT) and
aspartate aminotransferase (AST) in patients with CHC and healthy
controls were detected using the standard automatic biochemistry
analyzer (AU5400; Olympus Corporation, Tokyo, Japan).
Patient enrollment
Between January 2012 and Feburary 2012, 43
consecutive patients with CHC and 22 healthy volunteers at the
First Affiliated Hospital of Chongqing Medical University were
recruited. The inclusion criteria for patients with biliary calculi
were based on the newly developed universal definition of biliary
calculi. Briefly, the patients with biliary calculi were clinically
diagnosed by biochemical markers, acute right upper quadrant
abdominal colicky pain and detection of calculi by sonography or
cholecystography. A total of 28 healthy volunteers with normal
liver function and no history of hepatobiliary disease were
recruited as non-biliary calculi controls.
Serum miRNA extraction and stem-loop
reverse transcription-quantitative PCR (RT-qPCR)
Using the mirVana PARIS miRNA isolation kit (Ambion
Life Technologies, Carlsbad, CA, USA), total RNA enriched with
miRNAs was extracted from the serum according to the manufacturer’s
instructions. RT-qPCR was conducted in order to determine the
expression levels of miR-196a. miRNAs were quantified through the
TaqMan miRNA RT-qPCR assay according to the manufacturer’s
instructions (Applied BioSystems Life Technologies, Foster City,
CA, USA). Briefly, RT-qPCR amplification was performed with
gene-specific forward primer and a reverse primer (Applied
Biosystems Life Technologies) along with a probe in an ABI Prizm
7500 PCR machine (Applied Biosystems Life Technologies), preceded
by first-strand cDNA synthesis with 10 ng RNA and
miRNA-196a-specific, stem-loop primer, or U6 stem-loop primer, a
control endogenous miRNA (Applied Biosystems Life Technologies).
The reverse-transcribed primers were designed as follows: miR-196a,
5′-GTCAGAAGGAATGATGCACAGCCAACAACA-3′; and U6:
5′-AACGCTTCACGAATTTGCGT-3′. The PCR primers were as follows: Mature
miR-196a, forward 5′-CGTCAGAAGGAATGATGCACAG-3′, and reverse
5′-ACCTGCGTAGGTAGTTTCATGT-3′; and U6, forward
5′-CTCGCTTCGGCAGCACA-3′, and reverse
5′-AACGCTTCACGAATTTGCGT-3′.
Relative miRNA expression was calculated
from experiments in triplicate following normalization to those for
U6 RNA
Relative miR-196a production, reported as
2‒∆∆Ct (Ct represents the threshold cycle), was
determined by the ∆Ct method. Differences in miR-196a concentration
between the two groups were expressed as fold changes.
Statistical analysis
Values are presented as the mean ± standard
deviation unless otherwise indicated. Spearman correlation
analysis, the Mann-Whitney U test, Student’s t-test, or the
χ2 test was conducted for between-group comparisons as
appropriate. The receiver operating characteristic (ROC) curves
were established for discriminating patients with CHC from the
normal controls. Two-tailed P<0.05 was considered to indicate a
statistically significant difference. All statistical calculations
were performed using SAS software, version 9.1.3 (SAS Institute,
Marlow, UK) and SPSS software, version 17.0 (SPSS, Inc., Chicago,
IL, USA).
Results
HepG2-HCV and HepG2-control groups
exhibit differences in miRNA expression profiles
A total of six differentially expressed miRNAs, with
a fold-difference ≥1.5 and P≤0.05, were identified between the
HepG2-HCV and HepG2-control cells following miRNA microarray
analysis (Fig. 1). Among these
miRNAs, miR-29a, miR146a, miR-149, miR-221 and miR-222 were
identified to be upregulated, while miR-196a was downregulated by
the overexpression of the HCV core protein (Table I).
 | Table IDifferentially expressed miRNA
profiles between HepG2-HCV and HepG2-control cells. |
Table I
Differentially expressed miRNA
profiles between HepG2-HCV and HepG2-control cells.
| Expression | miRNAs | Fold change |
|---|
| miR-29 | 1.6 |
| miR-146a | 2.0 |
| miR-149 | 1.8 |
| miR-221 | 1.8 |
| miR-222 | 1.5 |
| Downregulated | miR-196a | 1.9 |
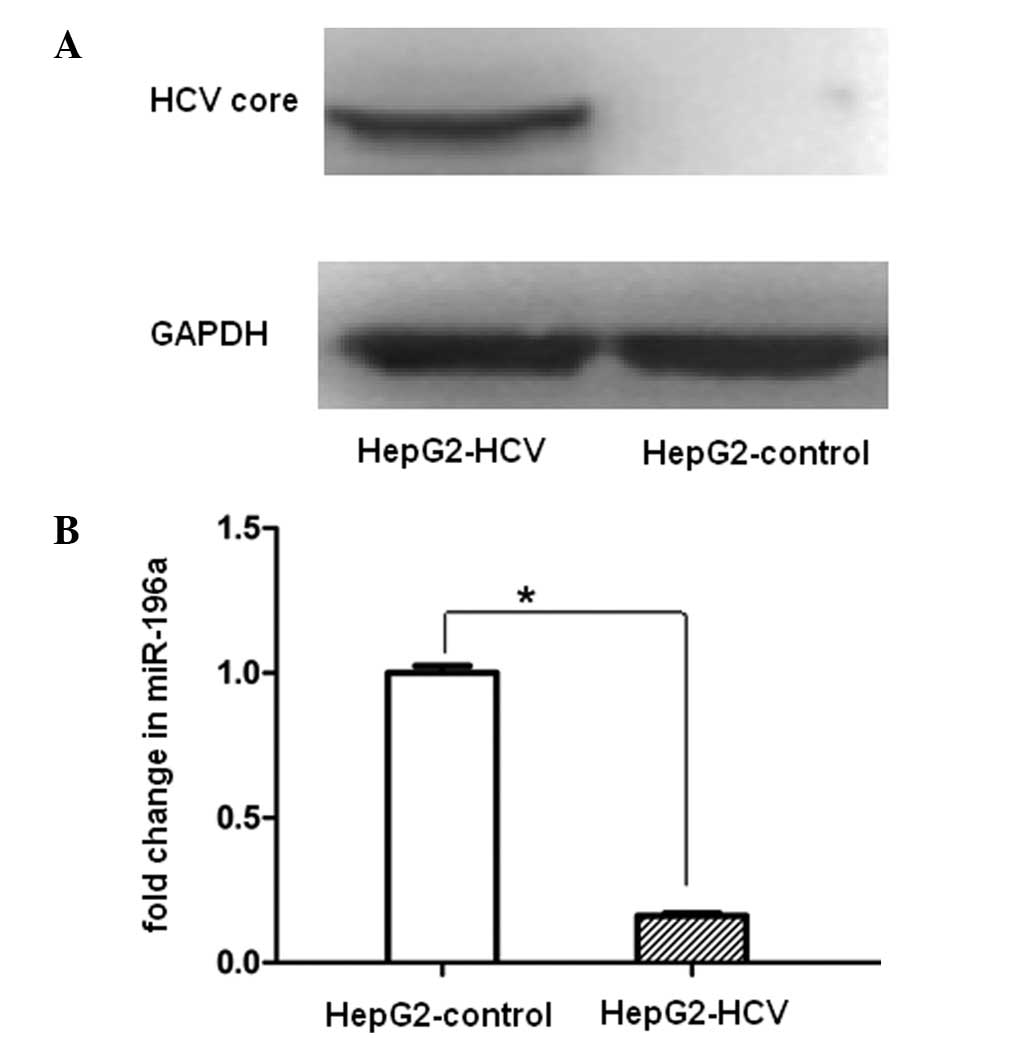
miR-196a is significantly downregulated
in the Ad-HCV infection group
To investigate whether the miR-196a expression
levels were affected by HCV core overexpression, HepG2 cells were
infected with Ad-HCV core. miR-196a was significantly downregulated
in HepG2-HCV cells as compared with that in the HepG2-control
following efficient expression of the HCV core protein at 48 h
(Fig. 2).
Serum miR-196a is significantly reduced
in patients with CHC and is diagnostically valuable for CHC
In order to investigate the clinical implications of
aberrant miR-196a expression and the use of circulating miR-196 in
the diagnosis and management of CHC, sera from 43 patients with CHC
and 22 healthy volunteers were collected for biomarker validation.
Between-group comparisons of the general clinical characteristics
demonstrated that there were no significant differences in the
gender ratio and mean age, but significant differences in ALT, AST
and HCV-RNA (Table II).
 | Table IIClinical characteristics of the
healthy control and chronic hepatitis C group. |
Table II
Clinical characteristics of the
healthy control and chronic hepatitis C group.
| Characteristic | Healthy control
group
(n=22) | Chronic hepatitis C
group
(n=43) | P-value |
|---|
| Age (mean ± standard
deviation) | 36.8±9.7 | 42.0±9.4 | <0.05 |
| Male, n (%) | 11 (50.0) | 27 (62.8) | >0.05 |
| Female, n (%) | 11 (50.0) | 16 (37.2) | |
| Alanine
aminotransferase (U/l) | 9.9±4.9 | 111.7±107.8 | <0.001 |
| Aspartate
aminotransferase (U/l) | 11.0±5.5 | 73.0±55.8 | <0.001 |
| Hepatitis C virus-RNA
(copies/ml) |
<1.0×103 |
>1.0×103 | <0.001 |
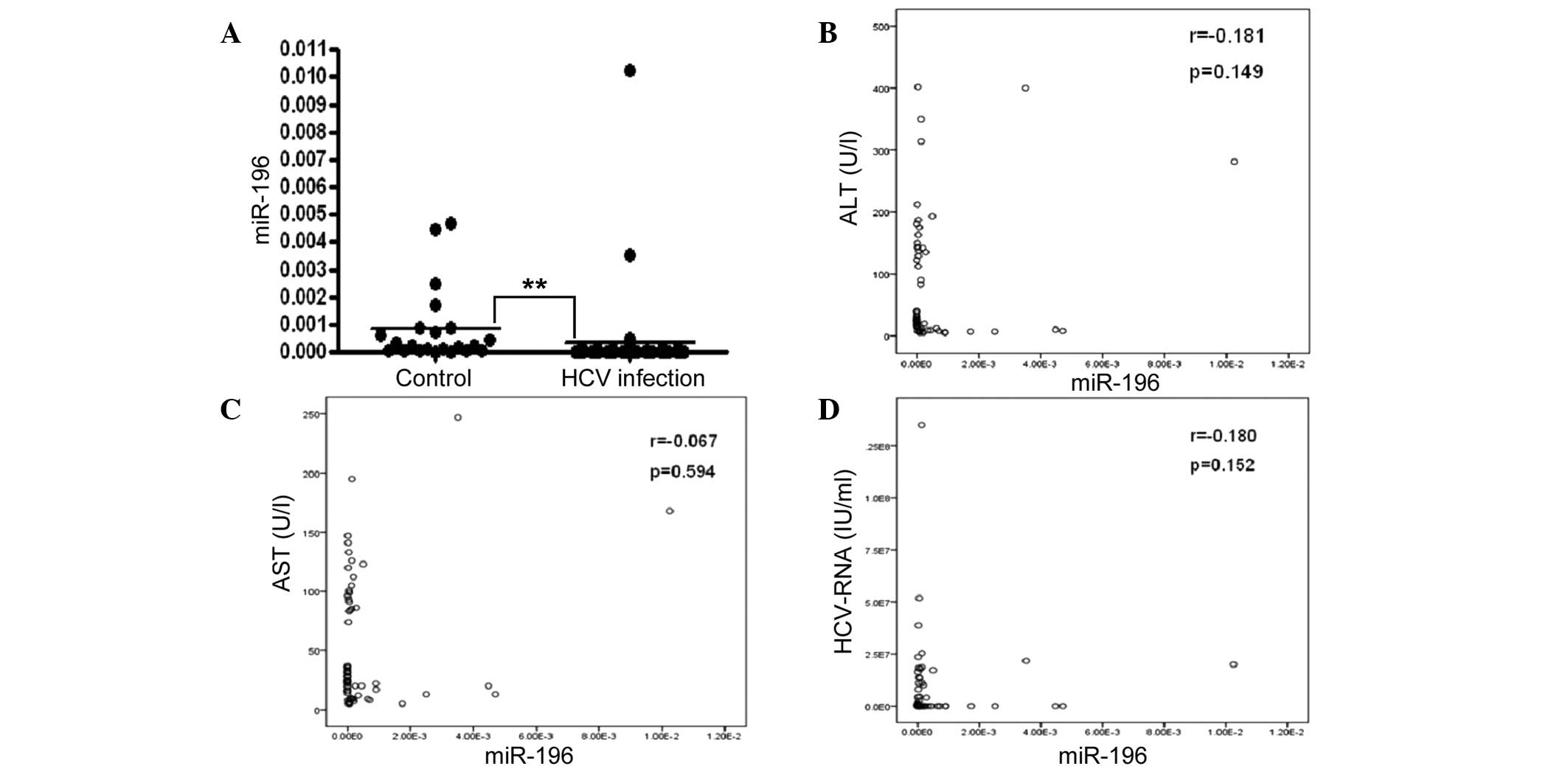
Circulating miR-196a was observed to be
significantly lower in the CHC group as compared with that in the
control group (P<0.001; Fig.
3A). Investigation of the possible correlation between
circulating miR-196a levels and the liver injury degree identified
no correlation between serum miR-196a and ALT/AST (Fig. 3B and C). Nor was a correlation
observed between miR-196a and HCV-RNA (Fig. 3D).
To further investigate the characteristics of
miR-196a as a potential biomarker of CHC, ROC curve analysis was
performed. Analysis of the ROC curves for serum miR-196a
demonstrated an AUC (area under the ROC curve) of 0.849 (95%CI:
0.756–0.941; P<0.001) with 81.8% sensitivity and 76.7%
specificity in discriminating chronic HCV infection from healthy
controls at a cut-off value of 6.115×10‒5 (Fig. 4). This suggested diagnostic value
of circulating miR-196a in CHC.
Discussion
Due to the absence of reliable and predictive
markers for the early diagnosis of HCV infection, treatment for CHC
is often delayed. Though HCV viral load analysis has impacted the
evaluation of the response likelihood of patients to therapy with
PEGylated IFN and ribavirin (18),
viral load monitoring is unable to assess the severity of disease
or risk of progression, as serum HCV RNA levels remain stable for
up to four years (19). The
present study confirmed that HCV core protein significantly
downregulated miR-196a expression in HepG2 cells. Furthermore, the
clinical implications of aberrant miR-196a expression and the use
of circulating miR-196 in the diagnosis and management of CHC was
validated by the fact that serum miR-196a levels were significantly
reduced in patients with CHC. Finally, serum miR-196a levels were
identified to be diagnostically valuable for CHC by producing an
AUC of 0.849 (95%CI: 0.756–0.941; P<0.001) with 81.8%
sensitivity and 76.7% specificity in discriminating CHC from
healthy controls at a cut-off value of 6.115×10‒5. These
results indicated the potential for use of circulating miR-196a as
a sensitive and informative biomarker for CHC. However, no
correlations were observed between the expression levels of
miR-196a, HCV viral load and ALT status.
miR-196 has been previously demonstrated to have
critical roles in normal development (20–22)
and in the pathogenesis of human malignancy (23–26),
immunology, inflammation and virus defense (12,27,28),
which has led to various studies attempting to decode its
functions. The present study suggested serum miR-196a as a novel
biomarker for CHC while profiling miRNAs in HCV core
protein-overexpressing HepG2 cells. The observation that the serum
miR-196a was relatively low in patients with CHC, but may be easily
detected in serum from healthy controls demonstrated for the first
time that monitoring of circulating miR-196a may also be applied in
clinical CHC diagnosis. ROC analysis identified that miR-196a may
be a sensitive, specific and practical clinical diagnostic
biomarker for CHC.
Although the present study had a small sample size,
it provided the first clinical evidence of the use of circulating
miR-196a as a biomarker of CHC, to the best of our knowledge.
However, further experiments with a larger sample size are required
to extensively evaluate the potential of miR-196a as a practical
biomarker. Circulating miRNAs are becoming attractive biomarker
candidates and are increasingly used in the prevention, diagnosis,
prognosis and therapeutic monitoring of various human diseases
(29). By demonstrating that
circulating miRNA levels returned to baseline levels following
tumorectomy, chemotherapy, acute myocardial infarction recovery and
other medical interventions, circulating miRNAs are proving to be
promising biomarkers for monitoring therapeutic effects (15). Thus it would be beneficial to
monitor the dynamic alterations in plasma miR-196a levels during
IFN treatment for CHC. In addition, various previous studies have
compared circulating miRNA biomarkers to existing markers and
demonstrated a strong correlation in miRNA expression and current
marker identification (30–32).
Furthermore, Resnick et al (33) and Zhu et al (34) reported that coupled with additional
established markers, circulating miRNAs demonstrate a greater
sensitivity than either used alone. Thus, serum miRNA biomarkers in
combination with other established biomarkers may provide
significant advantages in early diagnosis and prognosis prediction.
Therefore, it will be valuable to investigate combined miR-196a and
HCV-RNA detection in the assessment of disease severity and
progression risk during CHC.
In conclusion, circulating miR-196a was
significantly reduced in patients with CHC, potentially via reduced
release of miR-196a from HCV-infected hepatocytes. Thus, the
presence of reduced circulating miR-196a may be a novel sensitive
and specific biomarker for early detection of CHC in humans.
Acknowledgments
The authors would like to thank Dr Shifeng Huang for
her help in editing the manuscript.
References
|
1
|
Eckman MH, Talal AH, Gordon SC, Schiff E
and Sherman KE: Cost-effectiveness of screening for chronic
hepatitis C infection in the United States. Clin Infect Dis.
56:1382–1393. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Saludes V, González V, Planas R, et al:
Tools for the diagnosis of hepatitis C virus infection and hepatic
fibrosis staging. World J Gastroenterol. 20:3431–3442. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sarrazin C, Wedemeyer H, Cloherty G, et
al: Importance of very early HCV RNA kinetics for prediction of
treatment outcome of highly effective all oral direct acting
antiviral combination therapy. J Virol Methods. 214C:29–32.
2014.
|
|
4
|
Bartel DP: MicroRNAs: genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Eulalio A, Huntzinger E and Izaurralde E:
Getting to the root of miRNA-mediated gene silencing. Cell.
132:9–14. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Carthew RW and Sontheimer EJ: Origins and
Mechanisms of miRNAs and siRNAs. Cell. 136:642–655. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Williams AE: Functional aspects of animal
microRNAs. Cell Mol Life Sci. 65:545–562. 2008. View Article : Google Scholar
|
|
8
|
Jopling CL, Yi M, Lancaster AM, Lemon SM
and Sarnow P: Modulation of hepatitis C virus RNA abundance by a
liver-specific MicroRNA. Science. 309:1577–1581. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bandyopadhyay S, Friedman RC, Marquez RT,
et al: Hepatitis C virus infection and hepatic stellate cell
activation downregulate miR-29: miR-29 overexpression reduces
hepatitis C viral abundance in culture. J Infect Dis.
203:1753–1762. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ishida H, Tatsumi T, Hosui A, et al:
Alterations in microRNA expression profile in HCV-infected hepatoma
cells: Involvement of miR-491 in regulation of HCV replication via
the PI3 kinase/Akt pathway. Biochem Biophys Res Commun. 412:92–97.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Henke JI, Goergen D, Zheng J, Song Y,
Schüttler CG, Fehr C, Jünemann C and Niepmann M: microRNA-122
stimulates translation of hepatitis C virus RNA. EMBO J.
27:3300–3310. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Pedersen IM, Cheng G, Wieland S, Volinia
S, Croce CM, Chisari FV and David M: Interferon modulation of
cellular microRNAs as an antiviral mechanism. Nature. 449:919–922.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hou W, Tian Q, Zheng J and Bonkovsky HL:
MicroRNA-196 represses Bach1 protein and hepatitis C virus gene
expression in human hepatoma cells expressing hepatitis C viral
proteins. Hepatology. 51:1494–1504. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sarasin-Filipowicz M, Krol J, Markiewicz
I, Heim MH and Filipowicz W: Decreased levels of microRNA miR-122
in individuals with hepatitis C responding poorly to interferon
therapy. Nat Med. 15:31–33. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
Weiland M, Gao XH, Zhou L and Mi QS: Small
RNAs have a large impact: Circulating microRNAs as biomarkers for
human diseases. RNA Biol. 9:850–859. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Huang S, Xie Y, Yang P, Chen P and Zhang
L: HCV core protein-induced down-regulation of microRNA-152
promoted aberrant proliferation by regulating Wnt1 in HepG2 cells.
PLoS One. 9:e817302014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Jardim MJ, Dailey L, Silbajoris R and
Diaz-Sanchez D: Distinct microRNA expression in human airway cells
of asthmatic donors identifies a novel asthma-associated gene. Am J
Respir Cell Mol Biol. 47:536–542. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kowala-Piaskowska A, Słuzewski W,
Figlerowicz M and Mozer-Lisewska I: Factors influencing early
virological response in children with chronic hepatitis C treated
with pegylated interferon and ribavirin. Hepatol Res. 32:224–226.
2005.PubMed/NCBI
|
|
19
|
Hollingsworth RC, Sillekens P, van Deursen
P, Neal KR and Irving WL: Serum HCV RNA levels assessed by
quantitative NASBA: stability of viral load over time, and lack of
correlation with liver disease. The Trent HCV Study Group. J
Hepatol. 25:301–306. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hornstein E, Mansfield JH, Yekta S, Hu JK,
Harfe BD, McManus MT, Baskerville S, Bartel DP and Tabin CJ: The
microRNA miR-196 acts upstream of Hoxb8 and Shh in limb
development. Nature. 438:671–674. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ronshaugen M, Biemar F, Piel J, Levine M
and Lai EC: The Drosophila microRNA iab-4 causes a dominant
homeotic transformation of halteres to wings. Genes Dev.
19:2947–2952. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Qiu R, Liu Y, Wu JY, Liu K, Mo W and He R:
Misexpression of miR-196a induces eye anomaly in Xenopus laevis.
Brain Res Bull. 79:26–31. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Bloomston M, Frankel WL, Petrocca F,
Volinia S, Alder H, Hagan JP, Liu CG, Bhatt D, Taccioli C and Croce
CM: MicroRNA expression patterns to differentiate pancreatic
adenocarcinoma from normal pancreas and chronic pancreatitis. JAMA.
297:1901–1908. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Schotte D, Chau JC, Sylvester G, Liu G,
Chen C, van der Velden VH, Broekhuis MJ, Peters TC, Pieters R and
den Boer ML: Identification of new microRNA genes and aberrant
microRNA profiles in childhood acute lymphoblastic leukemia.
Leukemia. 23:313–322. 2009. View Article : Google Scholar
|
|
25
|
Maru DM, Singh RR, Hannah C, et al:
MicroRNA-196a is a potential marker of progression during Barrett’s
metaplasia-dysplasia-invasive adenocarcinoma sequence in esophagus.
Am J Pathol. 174:1940–1948. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Schimanski CC, Frerichs K, Rahman F,
Berger M, Lang H, Galle PR, Moehler M and Gockel I: High miR-196a
levels promote the oncogenic phenotype of colorectal cancer cells.
World J Gastroenterol. 15:2089–2096. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ye L, Wang X, Wang S, Wang Y, Song L, Hou
W, Zhou L, Li H and Ho W: CD56+ T cells inhibit hepatitis C virus
replication in human hepatocytes. Hepatology. 49:753–762. 2009.
View Article : Google Scholar :
|
|
28
|
Sonkoly E, Ståhle M and Pivarcsi A:
MicroRNAs and immunity: Novel players in the regulation of normal
immune function and inflammation. Semin Cancer Biol. 18:131–140.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Witwer KW: Circulating microRNA biomarker
studies: Pitfalls and potential solutions. Clin Chem. 61:56–63.
2015. View Article : Google Scholar
|
|
30
|
Zhong J, He Y, Chen W, et al: Circulating
microRNA-19a as a potential novel biomarker for diagnosis of acute
myocardial infarction. Int J Mol Sci. 15:20355–20364. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Shifeng H, Danni W, Pu C, et al:
Circulating liver-specific miR-122 as a novel potential biomarker
for diagnosis of cholestatic liver injury. PLoS One. 8:e731332013.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Li H, Wang Z, Fu Q and Zhang J: Plasma
miRNA levels correlate with sensitivity to bone mineral density in
postmenopausal osteoporosis patients. Biomarkers. 19:553–556. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Resnick KE, Alder H, Hagan JP, Richardson
DL, Croce CM and Cohn DE: The detection of differentially expressed
microRNAs from the serum of ovarian cancer patients using a novel
real-time PCR platform. Gynecol Oncol. 112:55–59. 2009. View Article : Google Scholar
|
|
34
|
Zhu W, Qin W, Atasoy U and Sauter ER:
Circulating microRNAs in breast cancer and healthy subjects. BMC
Res Notes. 2:892009. View Article : Google Scholar : PubMed/NCBI
|