Introduction
Preeclampsia (PE) is a vascular disorder, which
presents with hypertension and proteinuria during pregnancy. It is
a consequence of a number of pathophysiological processes,
including endothelial dysfunction and systemic inflammation. It is
a key risk factor for maternal and fetal morbidity and mortality
worldwide (1–3). Although the exact mechanisms
underlying the development of PE remain unclear, disorders of
maternal tissue, and maternal obesity and insulin resistance are
likely to be involved. Pathological manifestations associated with
this condition include poor placentation, shallow placental
invasion and abnormal angiogenesis. (4–7).
MicroRNAs (miRNAs) are single-stranded non-coding
RNAs (ncRNAs) composed of 18–24 nucleotides. They contribute to
numerous biological regulatory processes, such as tumorigenesis,
cell proliferation, cell differentiation and apoptosis, by inducing
the silencing of target messenger RNAs (mRNAs) (8–11).
During the formation of these molecules two types of intermediate
miRNA are created: Primary miRNA (pri-miRNA) and precursor miRNA
(pre-miRNA). Pri-miRNA, a long transcript from the 3′-untranslated
region, is cleaved by the Drosha enzyme to form pre-miRNA in the
nucleus. The cytoplasmic enzyme, Dicer, then processes pre-miRNA
into mature miRNA. The RNA-induced silencing complex is then
generated by the miRNA and catalyzes cleavage of a single
phosphodiester bond on the mRNA target (12–14).
Differentially expressed miRNAs are involved in certain human
diseases and may have a use as biomarkers of these conditions
(6,15–18).
For example, upregulated miR-155 is known to be a risk factor for
PE, via its regulation of cysteine-rich angiogenic inducer 61
(19).
Recently, a number of studies have suggested that
differentially expressed miRNAs may be involved in the development
of PE, since the biological processes involving these miRNAs have
been demonstrated to be similar to those involved in PE (20–22).
miRNAs may be useful as biomarkers for the diagnosis of PE.
However, the results from microarray studies are inconsistent, and
thus further investigation is required in order to reliably use
this method of analysis in clinical practice. For example, miR-182
exhibited different levels of expression levels in two different
studies (21,22). Therefore in the present study,
sequencing technology was used to detect the differentially, highly
and consistently expressed miRNAs that may be associated with the
development of PE. miRNA expression was measured in the plasma and
placenta of patients with mild and severe PE. Based on the results
of these experiments, a genome-wide screen for the deregulated
miRNAs was conducted. Simultaneously, the miRNA gene clusters or
families to which these miRNAs belong were defined. Functional
enrichment analyses were conducted in order to predict the pathways
involved in PE and the interaction between the target mRNAs.
Materials and methods
Sample collection and small RNA
sequencing
Samples were obtained from five subjects who had
delivered by elective cesarean section. They comprised four
patients with PE and one subject with a pregnancy without
complications, who were recruited from Zhongda Hospital (Nanjing,
China). The study protocol was approved by the Research Ethics
Board of Zhongda Hospital. Written informed consent was obtained
prior to blood sample collection. Maternal plasma and placenta
samples were collected from a normal pregnant female and the
patients with PE. Two patients were diagnosed with mild PE (mPE
group) and two with severe PE (sPE group; Table I). TRIzol® (Invitrogen
Life Technologies, Carlsbad, CA, USA) was used to extract total
RNA. mirVana™ miRNA Isolation kit (Ambion Life Technologies,
Austin, TX, USA) was used to isolate small miRNA from total RNA. An
miRNA library was constructed according to the manufacturer’s
instructions for the use of SOLiD™ Small RNA Expression kit
(Invitrogen Life Technologies). SOLiD sequencing platform (Applied
Biosystems Life Technologies, Foster City, CA, USA) was used to
sequence miRNAs. The sequencing process was completed at the State
Key Laboratory of Bioelectronics, School of Biological Science and
Medical Engineering, Southeast University, (Nanjing, China).
 | Table IClinical information from four
patients with preeclampsia. |
Table I
Clinical information from four
patients with preeclampsia.
| Patient | Severity | Age (years) | Gender of
infants | Length of pregnancy
(days) |
|---|
| m1 | Mild | 27 | Male | 280 |
| m2 | Mild | 26 | Male | 283 |
| s1 | Severe | 34 | Female | 244 |
| s2 | Severe | 28 | Female | 244 |
Raw sequencing datasets were obtained from other
ncRNAs, including small nucleolar RNAs, transfer RNAs and ribosomal
RNAs. Remaining reads were mapped to the known human pre-miRNAs in
the miRBase database (Release 16.0, http://www.mirbase.org/) with Bowtie 0.12.7 (23). Regardless of the adaptor sequence,
only one mismatch was permitted.
Identification of differentially
expressed miRNAs and their targets
Raw data was normalized. The percentage of each
miRNA from a single sample was taken as its expression level.
miRNAs that were consistently expressed in all samples were
selected, as these were assumed to be deregulated in patients with
mild and severe preeclampsia. Differentially-and highly-expressed
miRNAs were identified. Highly expressed miRNA were defined as
those where the percentage of the expression levels in a single
sample was >0.02. When the difference of the logarithmic value
of the level of an miRNA between patients with PE and the subject
with a normal pregnancy was >1.2 or <-1.2, it was defined as
a deregulated miRNA. Differentially and highly expressed miRNAs in
the plasma and placenta were thus selected for further analysis.
Gene clusters or families of deregulated miRNAs were identified by
miRBase (23). Three datasets,
including miRanda, TargetScan and miRTarBase, were used to identify
the targets of deregulated miRNAs (24–26).
One target was found in at least two datasets. The use of more than
one dataset was employed in order to reduce the rate of false
positive results.
Functional enrichment analysis
The targets of common miRNAs were investigated by
functional enrichment analysis. mRNAs in which the frequency of
targeting by miRNAs was ≥2 were enriched by the Gene Ontology
Biological Process (GOBP) database and the Kyoto Encyclopedia of
Genes and Genomes (KEGG) database (27). P-values represented the probability
of the involvement of the pathways enriched by the genes. q values
represent the false discovery rate (28). P<0.05 was considered to indicate
a statistically significant difference. The targets identified by
this process may have a causal role in the development of PE.
Results
Deregulated miRNAs and their
families
The total number of miRNAs detected was 905. The
number of miRNAs consistently expressed in placenta and plasma were
734 and 269, respectively. Highly-expressed miRNAs were selected
from these groups in the placenta and plasma (159 and 109,
respectively). In the placenta, 71 differentially-expressed miRNAs
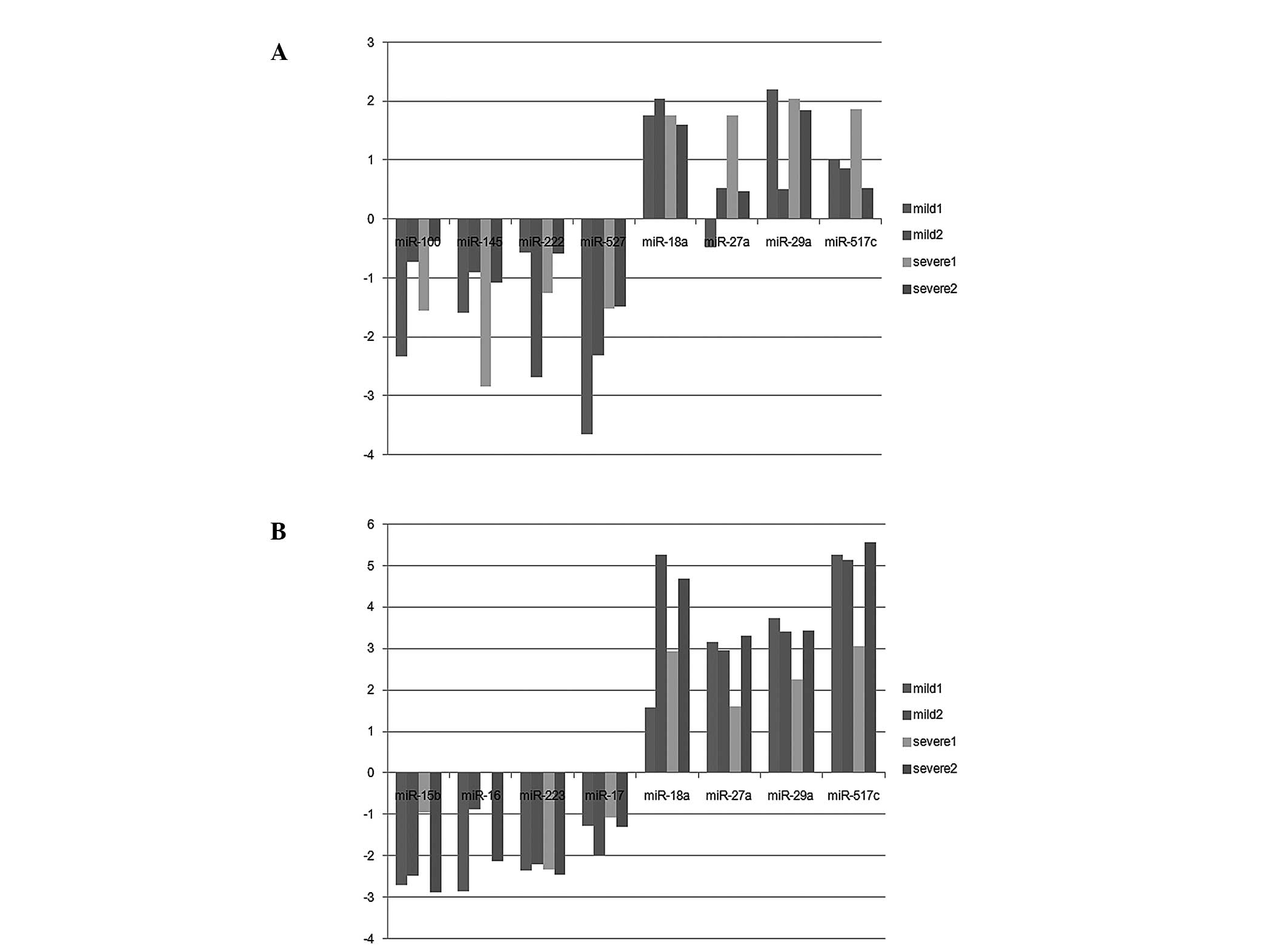
(26 upregulated and 45 downregulated) were identified (Fig. 1A). In the plasma, 94
differentially-expressed miRNAs (81 upregulated and 13
downregulated) were identified (Fig.
1B). Notably, the abnormally-expressed miRNAs observed in the
plasma and placenta were all upregulated, including miR-126,
miR-126*, miR-130a, miR-135b, miR-142-3p, miR-149, miR-188-5p,
miR-18a, miR-18b, miR-203, miR-205, miR-224, miR-27a, miR-29a,
miR-301a, miR-517c, miR-518-3p, miR-518e, miR-519d and miR-93
(Table II). Although expression
levels were abnormal, they differed between the placenta and plasma
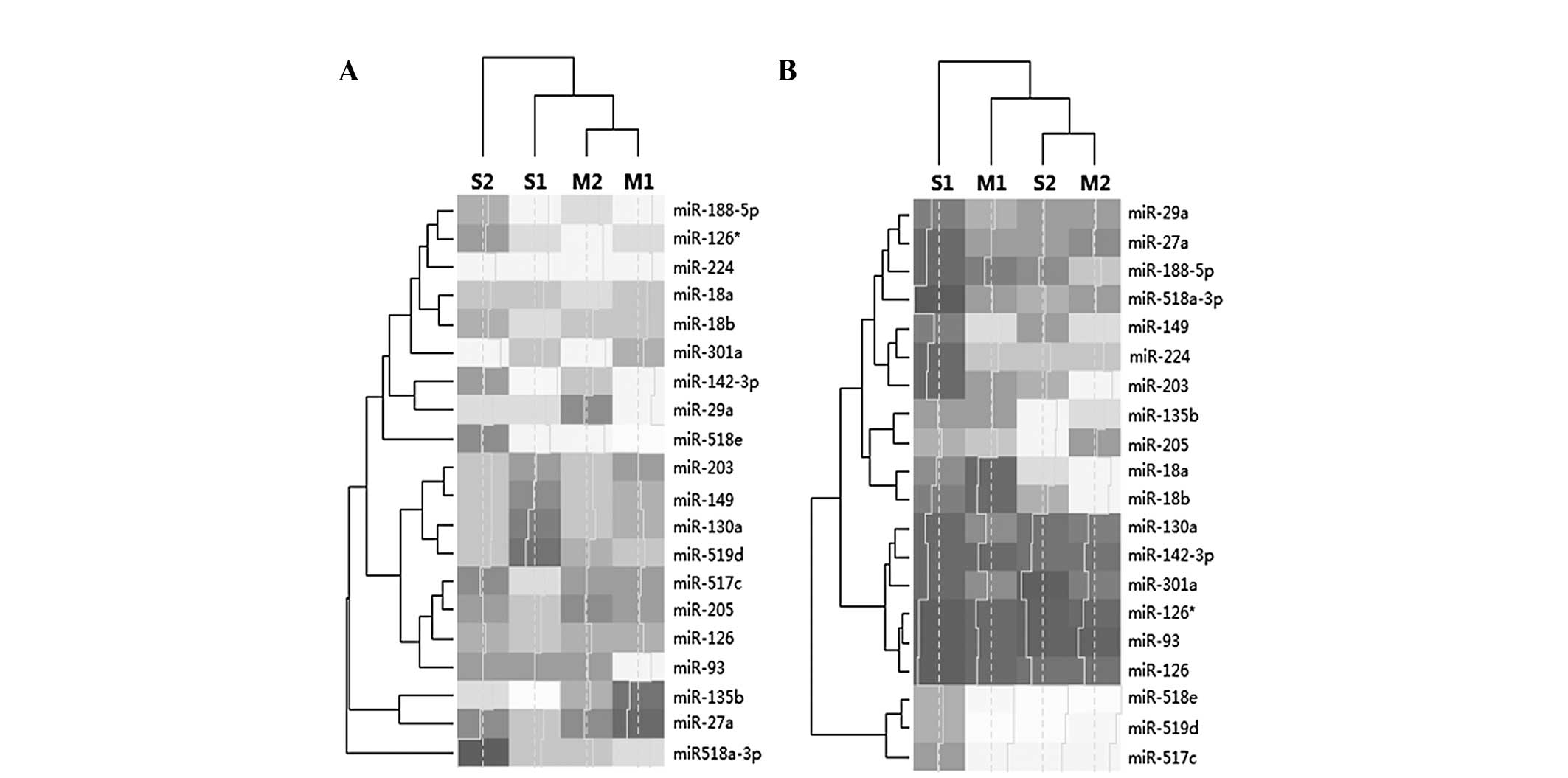
(Fig. 2). The miRNAs identified,
were members of 13 gene families or clusters, of which four
families contained two or more of these aberrantly expressed
miRNAs: miR 126, miR-126 and miR-126*; mir 17 family, miR-18a,
miR-18b and miR-93; miR 130 family, miR-130a and miR-301a; and miR
515 family, miR-517*, miR-518a-3p, miR-518e and miR-519d. miRNAs
involving the same family exhibited different expression levels
(Fig. 3). These deregulated miRNAs
were selected from 905 miRNAs following three experimental
processes; therefore they may have been the causal miRNAs and their
families may be the causal gene families.
 | Table IIAssociation of differentially
expressed miRNAs in placenta and plasma from patients with mild or
severe preelampsia. |
Table II
Association of differentially
expressed miRNAs in placenta and plasma from patients with mild or
severe preelampsia.
A, Placenta
|
|---|
|
|---|
| miRNA | Mild
| Severe
|
|---|
| log2FC | %a | log2FC | %a |
|---|
| miR-126 | 1.36 | 0.89 | 1.29 | 0.85 |
| miR-126* | 2.35 | 0.99 | 1.68 | 0.62 |
| miR-130a | 1.57 | 2.16 | 1.15 | 1.61 |
| miR-135b | 0.71 | 0.02 | 2.96 | 0.11 |
| miR-142 3p | 2.10 | 0.07 | 2.49 | 0.10 |
| miR-149 | 1.33 | 0.03 | 1.25 | 0.03 |
| miR-188 5p | 2.23 | 0.02 | 1.90 | 0.01 |
| miR-18a | 1.90 | 0.06 | 1.68 | 0.06 |
| miR-18b | 1.60 | 0.03 | 1.66 | 0.03 |
| miR-203 | 1.23 | 0.02 | 1.40 | 0.02 |
| miR-205 | 0.79 | 0.06 | 1.23 | 0.08 |
| miR-224 | 2.45 | 0.17 | 2.23 | 0.15 |
| miR-27a | 0.10 | 0.49 | 1.25 | 1.09 |
| miR-29a | 1.58 | 1.80 | 1.94 | 2.31 |
| miR-301a | 2.23 | 0.07 | 2.23 | 0.07 |
| miR-517c | 0.94 | 2.53 | 1.34 | 3.35 |
| miR-518a 3p | 1.78 | 0.11 | 0.61 | 0.05 |
| miR-518e | 3.26 | 0.96 | 1.84 | 0.36 |
| miR-519d | 1.30 | 6.52 | 0.94 | 5.09 |
| miR-93 | 1.65 | 0.05 | 0.81 | 0.03 |
|
| B, Plasma |
|
| miRNA | Mild
| Severe
|
| log2FC | %a | log2FC | %a |
|
| miR-126 | 1.67 | 0.35 | 1.18 | 0.25 |
| miR-126* | 1.51 | 0.37 | 0.81 | 0.23 |
| miR-130a | 2.51 | 0.45 | 1.66 | 0.25 |
| miR-135b | 4.05 | 0.05 | 4.34 | 0.06 |
| miR-142 3p | 1.74 | 0.07 | 1.73 | 0.07 |
| miR-149 | 4.52 | 0.06 | 3.03 | 0.02 |
| miR-188 5p | 3.35 | 0.03 | 2.20 | 0.01 |
| miR-18a | 4.36 | 0.34 | 4.05 | 0.27 |
| miR-18b | 4.05 | 0.18 | 3.26 | 0.11 |
| miR-203 | 4.81 | 0.04 | 2.96 | 0.01 |
| miR-205 | 3.75 | 0.13 | 4.35 | 0.20 |
| miR-224 | 4.21 | 0.10 | 3.41 | 0.06 |
| miR-27a | 3.06 | 0.54 | 2.69 | 0.42 |
| miR-29a | 3.57 | 1.51 | 2.96 | 0.98 |
| miR-301a | 2.50 | 0.07 | 1.19 | 0.03 |
| miR-517c | 5.19 | 2.11 | 4.79 | 1.60 |
| miR-518a 3p | 3.35 | 0.04 | 2.62 | 0.03 |
| miR-518e | 6.35 | 0.45 | 5.65 | 0.28 |
| miR-519d | 6.06 | 5.24 | 5.37 | 3.24 |
| miR-93 | 1.35 | 0.05 | 0.87 | 0.04 |
Target mRNAs and gene enrichment
analysis
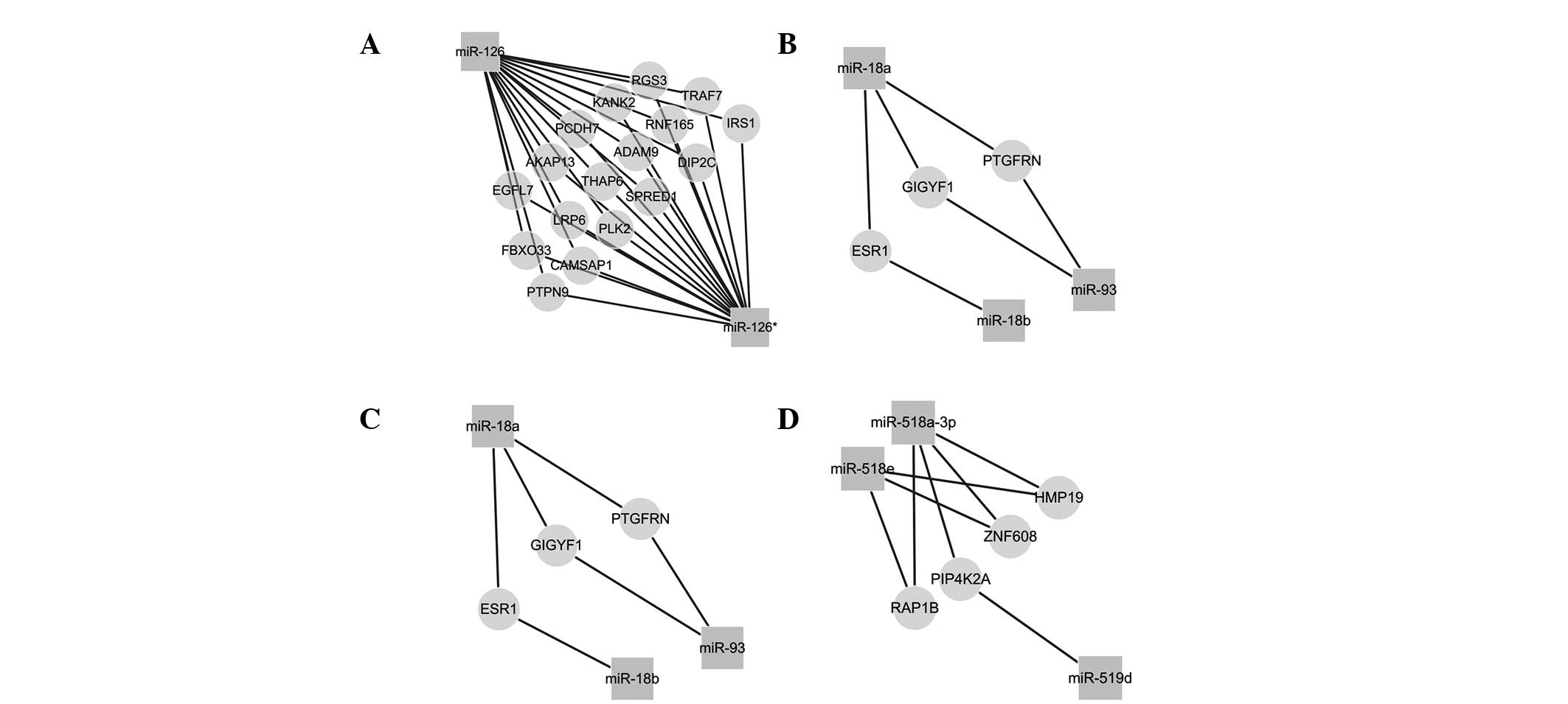
The targets of deregulated miRNAs were predicted
from three datasets, in order to increase the accuracy of this
prediction. 3,818 mRNAs were identified as targets of the 20
deregulated miRNAs. 1,215 mRNAs were found to be regulated by ≥2
miRNAs. Adaptor-associated protein 1 (AAK1), ankyrin repeat domain
2, F-Box protein 45, LOCR, nuclear factor I/B, neuropilin 2,
phosphatase and tensin homolog (PTEN) and ring finger protein, LIM
domain were found to be regulated by six or more deregulated miRNAs
(Table III). It is hypothesized
that these mRNAs were therefore associated with the development of
PE. Within the same gene family, these miRNAs may regulate similar
targets, and may thus be involved in similar biological processes
(Fig. 4). Furthermore, these mRNAs
were investigated using gene enrichment analysis by GOBP and KEGG
(Tables IV and V). From GOBP, these targets were enriched
into 1,337 pathways, amongst which 1,013 significant targets were
identified (P<0.05). Targets (200) were enriched in the
regulation of transcription (GOBPID: 0006355), which may be one of
the causal pathways in the pathogenesis of PE. In addition, 75
significant pathways were identified from 119 pathways from the
KEGG database. Focal adhesion, in which including 37 targets were
enriched, may be another pathway involved in this disease process.
Although the enrichment theories of GOBP and KEGG differed, the
pathways enriched by each of them were essential.
 | Table IIISummary of association of targets
(frequency≥6) and their miRNAs. |
Table III
Summary of association of targets
(frequency≥6) and their miRNAs.
| Target | Frequency | miRNA |
|---|
| AAK1 | 7 | miR-130a, miR-149,
miR-188-5p, miR-203, miR-205,miR-27a, miR-93 |
| ANKRD52 | 6 | miR-149, miR-18a,
miR-203, miR-224, miR-29a, miR-519d |
| FBXO45 | 6 | miR-135b,
miR-142-3p, miR-188-5p, miR-203, miR-27a, miR-29a |
| LCOR | 6 | miR-130a,
miR-142-3p, miR-203, miR-205, miR-224, miR-27a |
| NFIB | 6 | miR-142-3p,
miR-149, miR-203, miR-205, miR-224, miR-27a |
| NRP2 | 6 | miR-130a, miR-149,
miR-188-5p, miR-224, miR-27a, miR-93 |
| PTEN | 6 | miR-188-5p,
miR-18a, miR-205, miR-29a, miR-301a, miR-519d |
| RLIM | 6 | miR-130a, miR-203,
miR-205, miR-27a, miR-29a, miR-518e |
 | Table IVTop ten pathways of targets by
GOBP. |
Table IV
Top ten pathways of targets by
GOBP.
| GOBPID | Term | Count | P |
|---|
| GO:000635 | Regulation of
transcription, DNA dependent | 200 | 7.91 E-205 |
| GO:0006350 | Transcription | 164 | 1.56 E-146 |
| GO:0007275 | Development | 108 | 7.47 E-80 |
| GO:0007165 | Signal
transduction | 111 | 2.05 E-65 |
| GO:0045944 | Positive regulation
of transcription from RNA polymerase Ⅱ promoter | 41 | 6.42 E-55 |
| GO:0006468 | Protein amino acid
phosphorylation | 54 | 1.91 E-53 |
| GO:0003155 | Cell adhesion | 43 | 1.59 E-36 |
| GO:0007399 | Nervous system
development | 40 | 1.03 E-31 |
| GO:0016568 | Chromatin
modification | 26 | 1.49 E-31 |
| GO:0019941 |
Modification-dependent protein
catabolism | 33 | 3.49 E-29 |
 | Table VTop ten pathways of targets by
KEGG. |
Table V
Top ten pathways of targets by
KEGG.
| Pathway | Count | P | q |
|---|
| Focal adhesion | 37 | 1.74 E-26 | 2.61 E-24 |
| Colorectal
cancer | 23 | 2.32 E-21 | 4.97 E-20 |
| MAPK signaling
pathway | 36 | 6.71 E-21 | 1.01 E-19 |
| Wnt signaling
pathway | 28 | 1.27 E-20 | 1.74 E-19 |
| ErbB signaling
pathway | 21 | 2.24 E-18 | 1.98 E-17 |
| Axon guidance | 23 | 8.84 E 17 | 5.76 E-16 |
| Regulation of actin
cytoskeleton | 28 | 2.44 E-16 | 1.46 E-15 |
| Insulin signaling
pathway | 21 | 4.39 E-14 | 2.12 E-13 |
| Chronic myeloid
leukemia | 16 | 2.00 E-13 | 8.63 E-13 |
| Prostate
cancer | 17 | 2.01 E-13 | 8.63 E-13 |
Discussion
It was clear that the expression of miRNAs in the
placenta was higher than that in the plasma. In addition,
accounting for cluster analysis, the expression patterns in the
placentas of the two patients with mild PE were more similar that
those of the patients with severe PE. Therefore, the classification
of the placenta should be performed prior to the analysis of
plasma. However, miRNAs detected in plasma are more readily
available as a non-invasive biomarker for screening in PE (29,30).
It therefore appears logical to focus on those biomarkers that are
deregulated in the plasma and placenta, and which are thus
accessible and also discriminative.
Furthermore, a single miRNA may be expressed to a
different degree between patients and between different tissues in
the same patient. Common miRNAs may be utilized as non-invasive
biomarkers, particularly in placental diseases and PE (30,31).
miR-126 is generated from the EGFl 7 gene in mice and is known to
indirectly increase the actions of proangiongenic factors, vascular
endothelial growth factor (VEGF) and fibroblast growth factor by
diminishing Spred-1 (32–34). Increased levels of proangiogenic
factors may promote the development of PE (35). The deregulation of miR-126 in this
disease may suggest a high level of expression of certain
proangiogenic factors, and the consistency of expression further
indicates that miR-126 is important in the pathogenesis of PE.
Furthermore, certain deregulated miRNAs are related to the
development of hypoxic trophoblasts which may be essential in the
pathogenesis of PE (36,37). For example, when the trophoblast is
exposed to hypoxia, miR-205 depresses mediator of RNA polymerase II
transcription subunit 1 (MED1), improving placental development.
This indicates that it is essential in trophoblast injury (32,38).
In the present study, the consistently upregulated expression of
this miRNA in patients with mild and severe preeclampsia provides
further evidence for this hypothesis. Thus, the deregulated miRNAs
may influence the development and generation of PE through
regulation of their various targets, including VEGF and MED1.
The predicted targets and their downstream pathways
are therefore also likely to be important factors in the
development of PE. It is likely that the predicted targets are
associated with PE as well as other diseases of pregnancy. For
example, VEGFA, regulated by miR-126, miR-203, miR-205, miR-29a and
miR-93, contributes to the development and maintenance of the
glomerular filtration barrier (39). VEGF expression may reflect the
degree of hypoxia in the placenta, which may also be the case for
miR-126 (40). Therefore, miR-126
and VEGF may be a causal miRNA-mRNA module in the development of
PE. This association requires verification in future studies.
Although the enrichment aims of GOBP and KEGG differed, the
pathways enriched by each were essential for the improvement in the
diagnosis and treatment of PE. PTEN is regulated by six significant
miRNAs, and was identified as being involved in the pathways of
focal adhesion and prostate cancer by KEGG, and the pathway of
protein amino acid phosphorylation by GOBP. Over-expression of PTEN
induces soluble endoglin release from endothelial cells. This
triggers endothelial dysfunction, a characteristic feature of PE
(41). In an integrative view,
gene enrichment analysis associates disease mechanisms with
predicted targets. Notably, these enriched pathways are likely to
be associated with the immune response, although one target is
involved in different processes (21). The insulin signaling pathway may be
associated with PE, as a second messenger of insulin is known to be
involved in this disease (7).
In conclusion, the 20 deregulated miRNAs identified
in the current study may be useful as non invasive biomarkers. The
pathways enriched from their targets and their gene clusters or
families are likely to be involved in the development of PE.
Acknowledgments
This study was supported by the National Natural
Science Foundation of China (grant nos. 61301251, 81473070 and
81373102), the Research Found for the Doctoral Program of Higher
Education of China (no. 211323411002), the National Natural Science
Foundation of Jiangsu (no. BK20130885), the Research and Innovation
Project for College Graduates of Jiangsu Province (KYLX_0944), the
Natural Science Foundation of the Jiangsu Higher Education
Institutions (nos. 12KJB310003 and 13KJB330003) and the Priority
Academic Program Development of Jiangsu Higher Education
Institutions (PAPD).
References
|
1
|
Enquobahrie DA, Abetew DF, Sorensen TK,
Willoughby D, Chidambaram K and Williams MA: Placental microRNA
expression in pregnancies complicated by preeclampsia. Am J Obstet
Gynecol. 204:e12–e21. 2011.
|
|
2
|
Redman CW and Sargent IL: Latest advances
in understanding preeclampsia. Science. 308:1592–1594. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Grill S, Rusterholz C, Zanetti-Dällenbach
R, et al: Potential markers of preeclampsia- a review. Reprod Biol
Endocrinol. 7:702009. View Article : Google Scholar
|
|
4
|
Roberts JM and Cooper DW: Pathogenesis and
genetics of pre-eclampsia. Lancet. 357:53–56. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Broughton Pipkin F and Roberts JM:
Hypertension in pregnancy. J Hum Hypertens. 14:705–724. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Huppertz B: Placental origins of
preeclampsia: challenging the current hypothesis. Hypertension.
51:970–975. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Scioscia M, Gumaa K, Kunjara S, et al:
Insulin resistance in human preeclamptic placenta is mediated by
serine phosphorylation of insulin receptor substrate-1 and -2. J
Clin Endocrinol Metab. 91:709–717. 2006. View Article : Google Scholar
|
|
8
|
Yi C, Wang Q, Wang L, et al: MiR-663, a
microRNA targeting p21(WAF1/CIP1), promotes the proliferation and
tumorigenesis of nasopharyngeal carcinoma. Oncogene. 31:4421–4433.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yang Q, Lu J, Wang S, Li H, Ge Q and Lu Z:
Application of next-generation sequencing technology to profile the
circulating microRNAs in the serum of preeclampsia versus normal
pregnant women. Clin Chim Acta. 412:2167–2173. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Salim H, Akbar NS, Zong D, et al:
miRNA-214 modulates radiotherapy response of non-small cell lung
cancer cells through regulation of p38MAPK, apoptosis and
senescence. Br J Cancer. 107:1361–1373. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Guo L, Yang Q, Lu J, et al: A
comprehensive survey of miRNA repertoire and 3′ addition events in
the placentas of patients with pre eclampsia from high throughput
sequencing. PLoS One. 6:e210722011. View Article : Google Scholar
|
|
12
|
Yi R, Qin Y, Macara IG and Cullen BR:
Exportin-5 mediates the nuclear export of pre-microRNAs and short
hairpin RNAs. Genes Dev. 17:3011–3016. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lund E, Güttinger S, Calado A, Dahlberg JE
and Kutay U: Nuclear export of microRNA precursors. Science.
303:95–98. 2004. View Article : Google Scholar
|
|
14
|
Schwarz DS, Tomari Y and Zamore PD: The
RNA-induced silencing complex is a Mg2+ dependent
endonuclease. Curr Biol. 14:787–791. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lu J, Getz G, Miska EA, et al: MicroRNA
expression profiles classify human cancers. Nature. 435:834–838.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mitchell PS, Parkin RK, Kroh EM, et al:
Circulating microRNAs as stable blood-based markers for cancer
detection. Proc Natl Acad Sci USA. 105:10513–10518. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kotlabova K, Doucha J and Hromadnikova I:
Placental-specific microRNA in maternal circulation -
identification of appropriate pregnancy-associated microRNAs with
diagnostic potential. J Reprod Immunol. 89:185–191. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Guo L, Zhao Y, Yang S, Cai M, Wu Q and
Chen F: Genome-wide screen for aberrantly expressed miRNAs reveals
miRNA profile signature in breast cancer. Mol Biol Rep.
40:2175–2186. 2013. View Article : Google Scholar
|
|
19
|
Zhang Y, Diao Z, Su L, et al: MicroRNA-155
contributes to preeclampsia by down-regulating CYR61. Am J Obstet
Gynecol. 202:e1–e7. 2010.PubMed/NCBI
|
|
20
|
Hu Y, Li P, Hao S, Liu L, Zhao J and Hou
Y: Differential expression of microRNAs in the placentae of Chinese
patients with severe pre-eclampsia. Clin Chem Lab Med. 47:923–929.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Pineles BL, Romero R, Montenegro D, et al:
Distinct subsets of microRNAs are expressed differentially in the
human placentas of patients with preeclampsia. Am J Obstet Gynecol.
196:e1–e6. 2007.PubMed/NCBI
|
|
22
|
Zhu XM, Han T, Sargent IL, Yin GW and Yao
YQ: Differential expression profile of microRNAs in human placentas
from preeclamptic pregnancies vs normal pregnancies. Am J Obstet
Gynecol. 200:e1–e7. 2009.PubMed/NCBI
|
|
23
|
Griffiths-Jones S, Saini HK, van Dongen S
and Enright AJ: miRBase: tools for microRNA genomics. Nucleic Acids
Res. 36:D154–D158. 2008. View Article : Google Scholar :
|
|
24
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
John B, Enright AJ, Aravin A, Tuschl T,
Sander C and Marks DS: Human microRNA targets. PLoS Biol.
2:e3632004. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Hsu SD, Lin FM, Wu WY, et al: miRTarBase:
a database curates experimentally validated microRNA-target
interactions. Nucleic Acids Res. 39:D163–D169. 2011. View Article : Google Scholar
|
|
27
|
Ogata H, Goto S, Sato K, Fujibuchi W, Bono
H and Kanehisa M: KEGG: Kyoto encyclopedia of genes and genomes.
Nucleic Acids Res. 27:29–34. 1999. View Article : Google Scholar
|
|
28
|
Ashburner M, Ball CA, Blake JA, et al:
Gene ontology: tool for the unification of biology. The Gene
Ontology Consortium Nat Genet. 25:25–29. 2000.
|
|
29
|
Kosaka N, Iguchi H and Ochiya T:
Circulating microRNA in body fluid: a new potential biomarker for
cancer diagnosis and prognosis. Cancer Sci. 101:2087–2092. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Mouillet JF, Chu T, Hubel CA, Nelson DM,
Parks WT and Sadovsky Y: The levels of hypoxia-regulated microRNAs
in plasma of pregnant women with fetal growth restriction.
Placenta. 31:781–784. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Lázár L, Nagy B, Molvarec A, Szarka A and
Rigó J Jr: Role of hsa-miR-325 in the etiopathology of
preeclampsia. Mol Med Rep. 6:597–600. 2012.PubMed/NCBI
|
|
32
|
Fish JE, Santoro MM, Morton SU, et al:
miR-126 regulates angiogenic signaling and vascular integrity. Dev
Cell. 15:272–284. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wang S, Aurora AB, Johnson BA, et al: The
endothelial-specific microRNA miR-126 governs vascular integrity
and angiogenesis. Dev Cell. 15:261–271. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Gellhaus A, Schmidt M, Dunk C, Lye SJ and
Winterhager E: The circulating proangiogenic factors CYR61 (CCN1)
and NOV (CCN3) are significantly decreased in placentae and sera of
preeclamptic patients. Reprod Sci. 14(8 Suppl): 46–52. 2007.
View Article : Google Scholar
|
|
35
|
Morales Prieto DM and Markert UR:
MicroRNAs in pregnancy. J Reprod Immunol. 88:106–111. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Mouillet JF, Chu T, Nelson DM, Mishima T
and Sadovsky Y: MiR-205 silences MED1 in hypoxic primary human
trophoblasts. FASEB J. 24:2030–2039. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Maccani MA, Padbury JF and Marsit CJ:
miR-16 and miR-21 expression in the placenta is associated with
fetal growth. PLoS One. 6:e212102011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Muralimanoharan S, Maloyan A, Mele J, Guo
C, Myatt LG and Myatt L: MIR-210 modulates mitochondrial
respiration in placenta with preeclampsia. Placenta. 33:816–823.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Eremina V and Quaggin SE: The role of
VEGF-A in glomerular development and function. Curr Opin Nephrol
Hypertens. 13:9–15. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Tsatsaris V, Goffin F, Munaut C, et al:
Overexpression of the soluble vascular endothelial growth factor
receptor in preeclamptic patients: pathophysiological consequences.
J Clin Endocrinol Metab. 88:5555–5563. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Cudmore MJ, Ahmad S, Sissaoui S, et al:
Loss of Akt activity increases circulating soluble endoglin release
in preeclampsia: identification of inter-dependency between Akt-1
and heme oxygenase 1. Eur Heart J. 33:1150–1158. 2012. View Article : Google Scholar :
|