Introduction
Osteoporosis, characterized by the loss of bone mass
and strength, and the development of microarchitecture impairment
leading to fragility fractures, has become a significant clinical
problem in health care services dealing with aging populations
(1,2). The susceptibility to osteoporosis is
regulated by a variety of factors, such as genetic variants, age,
sex steroid production, lifestyle and environment (3–5).
A number of studies have investigated the
pathogenesis of osteoporosis at the molecular levels. Two
cytokines, including osteoprotegerin and receptor activator of
nuclear factor κB ligand, have been identified as important
regulators in the development of osteoporosis (2,6).
Members of the Wnt signaling pathway, such as low-density
lipoprotein receptor-related protein 5 (LRP5), Wnt3a, secreted
Frizzled-related protein 1 and sclerostin (SOST), have been
reported to be associated with variation in bone mineral density
(7). Additionally, Wnt signaling
may enhance osteoblast survival, and interact with parathyroid
hormone signaling and bone morphogenetic protein 2, leading to an
elevation in osteoblastogenesis (8–10).
The transcription factor, specificity protein 1 (Sp1), is
associated with a reduction in bone quality and the biomechanical
properties of bone (11). However,
the underlying etiology of osteoporosis is not yet comprehensively
understood and the identification of novel therapeutic targets for
osteoporosis is required.
Mesenchymal stem cells (MSCs) from bone marrow are
multipotent cells that are able to differentiate into multiple cell
lineages, including osteoblasts, adipocytes, fibroblasts and
chondrocytes (12,13). The implantation of MSCs has been
shown to be an effective and safe method by which to enhance bone
regeneration and repair in animal models for bone regeneration as
well as in clinical practice (14,15).
Gene-expression microarrays are a powerful tool with
high-throughput technology, which may be used to assess the
expression patterns of multiple genes simultaneously. Therefore,
gene expression microarray analysis of MSCs from patients with
osteoporosis, may provide novel insights into the mechanisms
underlying the pathogenesis of osteoporosis.
In the present study, gene expression profiles of
MSCs from patients with osteoporosis and controls were downloaded,
in order to identify differentially expressed genes (DEGs). The
screened DEGs were further analyzed using bioinformatics methods to
reveal osteoporosis-specific gene expression patterns. The aim was
to provide novel targets for the diagnosis and treatment of
osteoporosis.
Materials and methods
Samples and data preprocessing
The gene expression profile of GSE35958 (16) was downloaded from the National
Center of Biotechnology Information Gene Expression Omnibus (GEO,
http://www.ncbi.nlm.nih.gov/geo/),
including five samples of human MSCs from the femoral heads of
elderly patients with osteoporosis and four control bone marrow
samples from age-matched non-osteoporotic donors. The platform used
was GPL570 [HG-U133_Plus_2] Affymetrix Human Genome U133 Plus 2.0
Array (Affymetrix UK Ltd, High Wycombe, United Kingdom).
The downloaded data in CEL files was preprocessed
using the Affy package. Background correction and quartile data
normalization were performed using the robust multiarray average
algorithm (17). Probes without a
corresponding gene symbol were then filtered and the average value
of gene symbols with multiple probes was calculated. Finally, the
expression profile dataset, including 20,539 genes for the nine
samples, was obtained.
Screening DEGs
Student’s t-test was used to identify DEGs between
the osteoporosis and control samples. The Benjamini-Hochberg (BH)
procedure (18) was used to adjust
the raw P-values into false discovery rates (FDRs). The DEGs were
screened using cut-off criteria of FDR<0.1 and
|log2FC|>1.5.
Functional and pathway enrichment
analysis for DEGs
In order to identify biological functions associated
with the pathogenesis of osteoporosis, Gene Ontology (GO) (19) functional and Kyoto Encyclopedia of
Genes and Genomes (KEGG) (20)
pathway enrichment analyses were performed for the identified DEGs,
using the online tool of Database for Annotation, Visualization and
Integrated Discovery (DAVID) (21)
based on the method of Expression Analysis Systemic Explorer (EASE)
test (22). The enrichment
threshold was an EASE score of 0.1.
Construction of the protein-protein
interaction network
Following the acquisition of pathways in which the
DEGs with FDR<0.1 and |log2FC|>1.5 were markedly
enriched, a protein-protein interaction (PPI) network of the
significant pathways was constructed, based on the Human Protein
Reference Database (HPRD) (23).
DEGs, which may convey effective information regarding the
pathogenesis of osteoporosis in the constructed PPI network were
identified.
Construction of co-change network for
pathways
The co-change network for pathways was established
based on the method of cumulative hypergeometric probability
distribution (24). The
pathway-pathway interactions with P<0.01 were identified to
construct the co-change network for pathways. P-values were
calculated using the following formula:
Where N is the total number of protein-protein interactions
involved with DEGs, M is the number of protein-protein interactions
associated with DEGs in a pathway, n is the number of
protein-protein interactions involved with DEGs in other pathways
and k is the number of protein-protein interactions involved with
DEGs between the two pathways.
Establishment of transcriptional
regulatory network for DEGs
TRANSFAC (25) is a
database containing information on eukaryotic transcription
regulating DNA sequence elements, their genomic binding sites and
their DNA-binding profiles. The transcriptional regulatory network
for DEGs was constructed based on the TRANSFAC database.
Results
Identification of differentially
expressed genes
Student’s t-test and the BH procedure were used to
identify DEGs between the osteoporosis and control samples. A total
of 1,127 DEGs were identified, with the cut-off criteria of
FDR<0.1 and |log2FC|>1.5, including 554
upregulated and 573 downregulated DEGs.
Functional and pathway enrichment
analysis for DEGs
The screened DEGs were used for functional and
pathway enrichment analysis by DAVID. A total of 27 DEGs had
significant involvement in the hsa04510 pathway (focal adhesion;
FDR=0.0205) and 17 DEGs were significantly enriched in the hsa04142
pathway (lysosome; FDR= 0.0477).
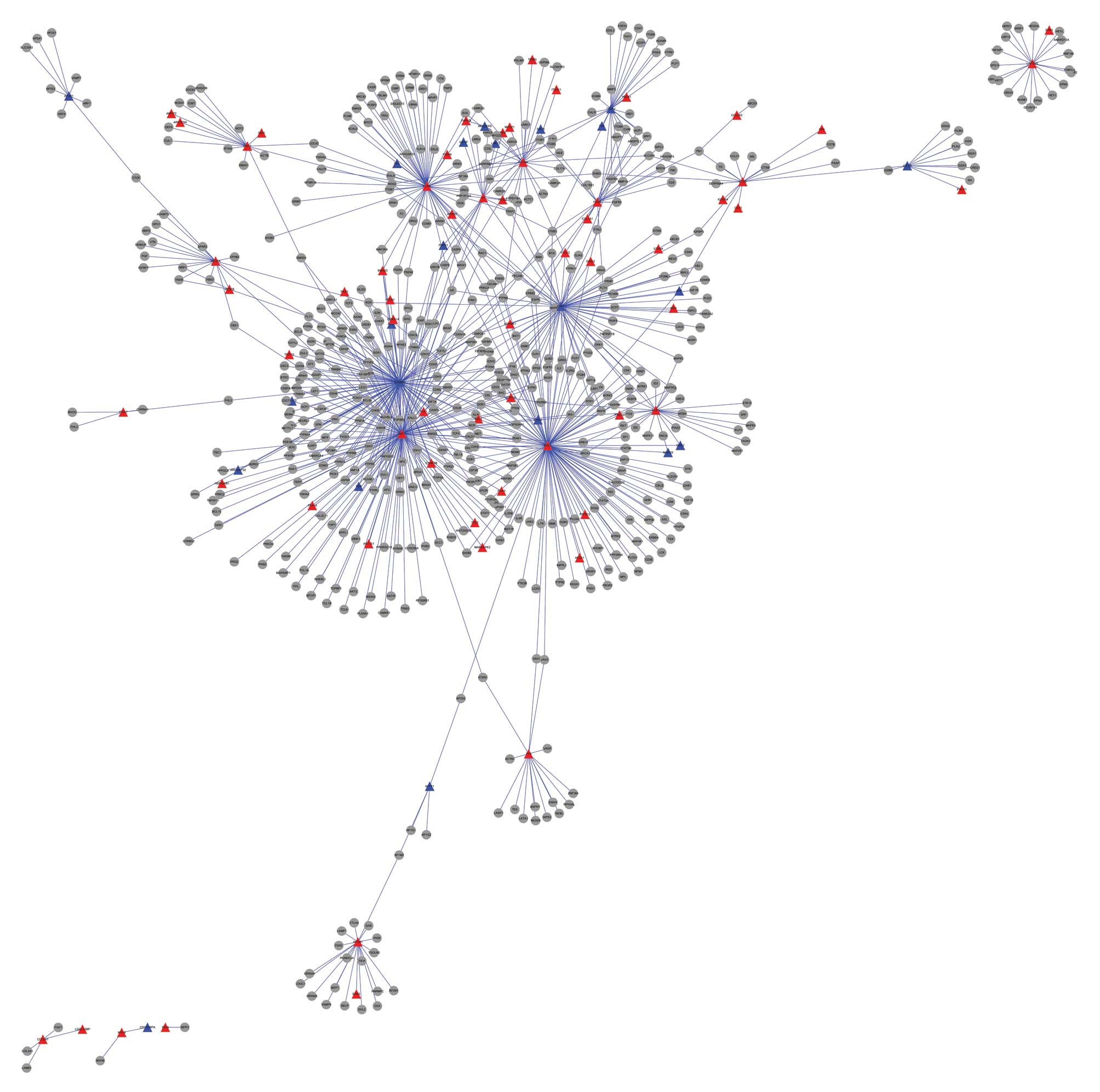
Protein-protein interaction network
construction
The protein-protein interactions for DEGs involved
in the two significant pathways, including focal adhesion and
lysosome, were identified, and were used to construct a PPI network
(Fig. 1). Table I shows DEGs with a degree >60 in
the constructed PPI network, including β-catenin (CTNNB1, 135),
SHC-transforming protein 1 (SHC1, 117), RAC-α
serine/thre-onine-protein kinase (AKT1, 117), caveolin 1 (CAV1, 73)
and filamin A (FLNA, 63). CTNNB1 and CAV1 were significantly
downregulated in the samples from the patients with osteoporosis,
while the other three genes were upregulated (Table I).
 | Table IFive differentially expressed genes
with degrees >60 in the constructed protein-protein interaction
network. |
Table I
Five differentially expressed genes
with degrees >60 in the constructed protein-protein interaction
network.
| Symbol | Gene ID | Degree | Type |
|---|
| CTNNB1 | 1499 | 135 | Downregulated |
| SHC1 | 6464 | 117 | Upregulated |
| AKT1 | 207 | 117 | Upregulated |
| CAV1 | 857 | 73 | Downregulated |
| FLNA | 2316 | 63 | Upregulated |
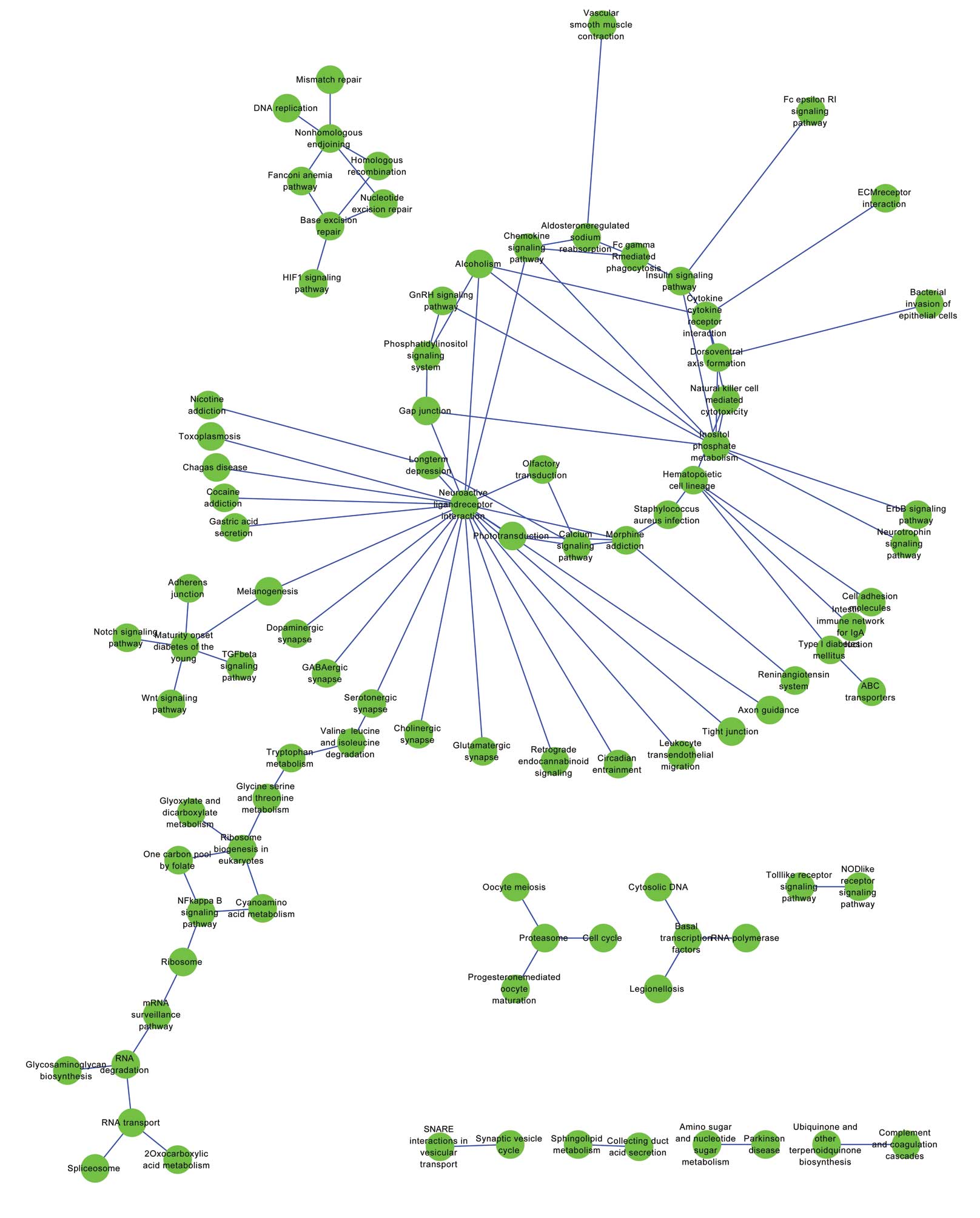
Establishment of a co-change network for
pathways
A total of 227 pathways were annotated for the PPI
network and the pathway-pathway interactions with P<0.01 were
identified in order to construct the co-change network for pathways
(Fig. 2). The co-change pathways
with degree ≥4 are shown in Table
II. The pathway with the degree of 22 in the constructed
co-change network was neuroactive ligand receptor interaction
(Table II). Other pathways were
inositol phosphate metabolism (degree, 9), cytokine receptor
interaction (degree, 5), hematopoietic cell lineage (degree, 5),
the calcium signaling pathway (degree, 4) and the chemokine
signaling pathway (degree, 4).
 | Table IITen pathways with degrees ≥4 in the
constructed co-change network for pathways. |
Table II
Ten pathways with degrees ≥4 in the
constructed co-change network for pathways.
| Pathway | Degree |
|---|
| Neuroactive ligand
receptor interaction | 22 |
| Inositol phosphate
metabolism | 9 |
| Nonhomologous end
joining | 5 |
| Cytokine receptor
interaction | 5 |
| Hematopoietic cell
lineage | 5 |
| Morphine
addiction | 5 |
| Maturity onset
diabetes of the young | 5 |
| Base excision
repair | 4 |
| Calcium signaling
pathway | 4 |
| Chemokine signaling
pathway | 4 |
Construction of a transcriptional
regulatory network for DEGs
A transcriptional regulatory network for DEGs was
constructed according to the information included in the TRANSFAC
database (Fig. 3). Genes with
degrees ≥9 in the transcriptional regulatory network are listed in
Table III. The nine genes with
high degrees in the constructed transcriptional regulatory network
were REL-associated protein (RELA), upstream stimulatory factor 1
(USF1), Sp1, Fos-related antigen 1 (FOSL1), cyclin-dependent kinase
inhibitor 1A (CDKN1A), upstream stimulatory factor 2 (USF2), ETS
domain-containing protein Elk-1 (ELK1), JUND and retinoic acid
receptor α (RARA), with degrees of 29, 27, 19, 18, 17, 13, 11, 11
and 9, respectively (Table III).
From the transcriptional regulatory network, Sp1 was shown to have
transcriptional regulatory associations with FOSL1, RELA and
CDKN1A.
 | Table IIINine genes with degrees ≥9 in the
constructed transcriptional regulatory network. |
Table III
Nine genes with degrees ≥9 in the
constructed transcriptional regulatory network.
| Symbol | Gene ID | Degree |
|---|
| RELA | 5970 | 29 |
| USF1 | 7391 | 27 |
| SP1 | 6667 | 19 |
| FOSL1 | 8061 | 18 |
| CDKN1A | 1026 | 17 |
| USF2 | 7392 | 13 |
| ELK1 | 2002 | 11 |
| JUND | 3727 | 11 |
| RARA | 5914 | 9 |
Discussion
The polymorphisms of a number of genes, including
vitamin D receptor, estrogen receptor α, estrogen receptor β, LRP5
and SOST, are associated with a risk of developing osteoporosis
(26,27). In the present study, the gene
expression profiles of hMSCs samples from elderly patients
suffering from osteoporosis and control samples were downloaded. A
total of 1,127 DEGs, including 554 upregulated and 573
downregulated DEGs, were screened. Functional and pathway
enrichment analyses revealed a significant involvement of DEGs in
the pathways of focal adhesion and lysosome. Focal adhesion kinase
may regulate the realignment of hMSCs, which is induced by
mechanical stretch (28). A number
of genes in the focal adhesion family have been reported as
candidate genes for osteoporosis (29). Therefore, the signaling pathway of
focal adhesion is likely to be important in the pathogenesis of
osteoporosis.
The DEGs that were involved in the two significantly
enriched pathways were used to construct a PPI network. The gene
with degree of 135 in the constructed protein-protein interaction
network was CTNNB1. Wnt/β-catenin signaling is involved in the
anabolic response to mechanical stimulation, and bone mass accrual
and maintenance. In addition, β-catenin has been shown to regulate
osteoblast survival and differentiation (10,30,31).
Ablation of β-catenin may promote the differentiation of osteoclast
precursors into bone-resorbing osteoclasts, ultimately leading to
osteoporosis (32). The results of
the current study also showed that CTNNB1 and CAV1 were
significantly downregulated in the osteoporosis samples, while
SHC1, AKT1 and FLNA were upregulated.
The pathway with the degree of 22 in the constructed
co-change network was neuroactive ligand receptor interaction. The
inositol phosphate metabolism, cytokine receptor interaction,
hematopoietic cell lineage, calcium signaling and chemokine
signaling pathways also had relatively high numbers of
interactions. It has been reported that mechanical loading may lead
to an increase in the intracellular calcium concentration in
osteoblasts, resulting in the activation of AKT, which is
responsible for osteoblast survival and proliferation (33). Certain chemokines are essential for
bone metabolism, such as osteopontin, which has been reported to be
involved in the pathogenesis of osteoporosis (34). Therefore, the pathway of
neuroactive ligand receptor interaction may be important in the
development of osteoporosis.
The genes with high degrees in the constructed
transcriptional regulatory network, were RELA, USF1, SP1, FOSL1,
CDKN1A, USF2, ELK1, JUND and RARA. Activation of liver X receptor
upregulates the expression of osteoclast/macrophage-related
markers, including USF1/2, which has the potential to inhibit the
differentiation of bone marrow-derived osteoclast precursors into
osteoclasts (35). Furthermore,
Sp1 had associations with the transcriptional regulation of FOSL1,
RELA and CDKN1A. The collagen type I α1 and Sp1 polymorphisms are
associated with reduced bone density and osteoporosis (36). Overexpression of Fos-related
antigen 1, encoded by FOSL1, may increase bone formation and
accelerate osteoblast differentiation in mice (37). Therefore, FOSL1, RELA and CDKN1A
may also be also involved in the pathogenesis of osteoporosis.
In conclusion, the significant DEGs identified in
the constructed PPI network and transcriptional regulatory network
may provide useful information on the pathogenesis of osteoporosis.
However, the present study did not analyze hMSCs from patients of
different ages and genders. Furthermore, the results of the study
require confirmation by experimental research. Therefore, the
molecular mechanism underlying the development of osteoporosis
demand further exploration.
Acknowledgments
This study was funded by Key Disciplines Group
Construction Project of Pudong Health Bureau of Shanghai
(PWZxq2014–9) and Shanghai Municipal Commission of Health and
Family Planning (grant no. 201440050).
References
|
1
|
Nguyen TV and Eisman JA: Genetic profiling
and individualized assessment of fracture risk. Nat Rev Endocrinol.
9:153–161. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Rachner TD, Khosla S and Hofbauer LC:
Osteoporosis: now and the future. Lancet. 377:1276–1287. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ralston SH and Uitterlinden AG: Genetics
of osteoporosis. Endocr Rev. 31:629–662. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Pietschmann P, Rauner M, Sipos W and
Kerschan-Schindl K: Osteoporosis: an age-related and
gender-specific disease - a mini-review. Gerontology. 55:3–12.
2009. View Article : Google Scholar
|
|
5
|
Seeman E: Bone quality: the material and
structural basis of bone strength. J Bone Miner Metab. 26:1–8.
2008. View Article : Google Scholar
|
|
6
|
Kong YY, Yoshida H, Sarosi I, et al: OPGL
is a key regulator of osteoclastogenesis, lymphocyte development
and lymph-node organogenesis. Nature. 397:315–323. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sims AM, Shephard N, Carter K, et al:
Genetic analyses in a sample of individuals with high or low BMD
shows association with multiple Wnt pathway genes. J Bone Miner
Res. 23:499–506. 2008. View Article : Google Scholar
|
|
8
|
Kramer I, Keller H, Leupin O and Kneissel
M: Does osteocytic SOST suppression mediate PTH bone anabolism?
Trends Endocrinol Metab. 21:237–244. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Rawadi G, Vayssière B, Dunn F, Baron R and
Roman-Roman S: BMP-2 controls alkaline phosphatase expression and
osteoblast mineralization by a Wnt autocrine loop. J Bone Miner
Res. 18:1842–1853. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Almeida M, Han L, Bellido T, Manolagas SC
and Kousteni S: Wnt proteins prevent apoptosis of both uncommitted
osteoblast progenitors and differentiated osteoblasts by
beta-catenin-dependent and -independent signaling cascades
involving Src/ERK and phosphatidylinositol 3-kinase/AKT. J Biol
Chem. 280:41342–41351. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Jin H, Stewart TL, Hof RV, Reid DM, Aspden
RM and Ralston S: A rare haplotype in the upstream regulatory
region of COL1A1 is associated with reduced bone quality and hip
fracture. J Bone Miner Res. 24:448–454. 2009. View Article : Google Scholar
|
|
12
|
Kilian KA, Bugarija B, Lahn BT and Mrksich
M: Geometric cues for directing the differentiation of mesenchymal
stem cells. Proc Natl Acad Sci USA. 107:4872–4877. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Valtieri M and Sorrentino A: The
mesenchymal stromal cell contribution to homeostasis. J Cell
Physiol. 217:296–300. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Griffin M, Iqbal SA and Bayat A: Exploring
the application of mesenchymal stem cells in bone repair and
regeneration. J Bone Joint Surg Br. 93:427–434. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Jones E and Yang X: Mesenchymal stem cells
and bone regeneration: current status. Injury. 42:562–568. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Benisch P, Schilling T, Klein-Hitpass L,
et al: The transcriptional profile of mesenchymal stem cell
populations in primary osteoporosis is distinct and shows
overexpression of osteogenic inhibitors. PLoS One. 7:e451422012.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Irizarry RA, Bolstad BM, Collin F, Cope
LM, Hobbs B and Speed TP: Summaries of affymetrix GeneChip probe
level data. Nucleic Acids Res. 31:e152003. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Benjamini Y and Hochberg Y: Controlling
the false discovery rate: a practical and powerful approach to
multiple testing. J Roy Statist Soc Ser B Stat (Methodological).
57:289–300. 1995.
|
|
19
|
Ashburner M, et al: Gene Ontology: tool
for the unification of biology. Nature genetics. 2000.25(1): 25–29.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kanehisa M and Goto S: KEGG: kyoto
encyclopedia of genes and genomes. Nucleic acids research.
2000.28(1): 27–30. View Article : Google Scholar
|
|
21
|
Da Wei Huang BTS and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nature protocols. 2008.4(1): 44–57.
View Article : Google Scholar
|
|
22
|
Hosack DA, Dennis G Jr, Sherman BT, Lane
HC and Lempicki RA: Identifying biological themes within lists of
genes with EASE. Genome Biol. 4:R702003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Stelzl U, Worm U, Lalowski M, et al: A
human protein-protein interaction network: a resource for
annotating the proteome. Cell. 122:957–968. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Przulj N, Wigle DA and Jurisica I:
Functional topology in a network of protein interactions.
Bioinformatics. 20:340–348. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wingender E, Dietze P, Karas H and Knüppel
R: TRANSFAC: a database on transcription factors and their DNA
binding sites. Nucleic Acids Res. 24:238–241. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li WF, Hou SX, Yu B, Li MM, Férec C and
Chen JM: Genetics of osteoporosis: accelerating pace in gene
identification and validation. Hum Genet. 127:249–285. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Jin H and Ralston SH: Regulatory
polymorphisms and osteoporosis. Gene Regulatory Sequences and Human
Disease. Springer; New York, NY: pp. p41–p54. 2012
|
|
28
|
Xu B, Song G and Ju Y: Effect of focal
adhesion kinase on the regulation of realignment and tenogenic
differentiation of human mesenchymal stem cells by mechanical
stretch. Connect Tissue Res. 52:373–379. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zintzaras E, Doxani C, Koufakis T,
Kastanis A, Rodopoulou P and Karachalios T: Synopsis and
meta-analysis of genetic association studies in osteoporosis for
the focal adhesion family genes: the CUMAGAS-OSTEOporosis
information system. BMC Med. 9:92011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Westendorf JJ, Kahler RA and Schroeder TM:
Wnt signaling in osteoblasts and bone diseases. Gene. 341:19–39.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Lau KH, Kapur S, Kesavan C and Baylink DJ:
Up-regulation of the Wnt, estrogen receptor, insulin-like growth
factor-I, and bone morphogenetic protein pathways in C57BL/6 J
osteoblasts as opposed to C3H/HeJ osteoblasts in part contributes
to the differential anabolic response to fluid shear. J Biol Chem.
281:9576–9588. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Otero K, Shinohara M, Zhao H, et al: TREM2
and β-catenin regulate bone homeostasis by controlling the rate of
osteoclasto-genesis. J Immunol. 188:2612–2621. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Rangaswami H, Schwappacher R, Tran T, et
al: Protein kinase G and focal adhesion kinase converge on
Src/Akt/β-catenin signaling module in osteoblast
mechanotransduction. J Biol Chem. 287:21509–21519. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Altintaş A, Saruhan-Direskeneli G, Benbir
G, Demir M and Purisa S: The role of osteopontin: a shared pathway
in the pathogenesis of multiple sclerosis and osteoporosis? J
Neurol Sci. 276:41–44. 2009. View Article : Google Scholar
|
|
35
|
Robertson Remen KM, Gustafsson JÅ and
Andersson G: The liver X receptor promotes macrophage
differentiation and suppresses osteoclast formation in mouse
RAW264.7 promy-elocytic leukemia cells exposed to bacterial
lipopolysaccharide. Biochem Biophys Res Commun. 430:375–380. 2013.
View Article : Google Scholar
|
|
36
|
Grant SF, Reid DM, Blake G, Herd R,
Fogelman I and Ralston SH: Reduced bone density and osteoporosis
associated with a polymorphic Sp1 binding site in the collagen type
I alpha 1 gene. Nat Genet. 14:203–205. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Jochum W, David JP, Elliott C, et al:
Increased bone formation and osteosclerosis in mice overexpressing
the transcription factor Fra-1. Nat Med. 6:980–984. 2000.
View Article : Google Scholar : PubMed/NCBI
|