|
1
|
Nguyen TV and Eisman JA: Genetic profiling
and individualized assessment of fracture risk. Nat Rev Endocrinol.
9:153–161. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Rachner TD, Khosla S and Hofbauer LC:
Osteoporosis: now and the future. Lancet. 377:1276–1287. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ralston SH and Uitterlinden AG: Genetics
of osteoporosis. Endocr Rev. 31:629–662. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
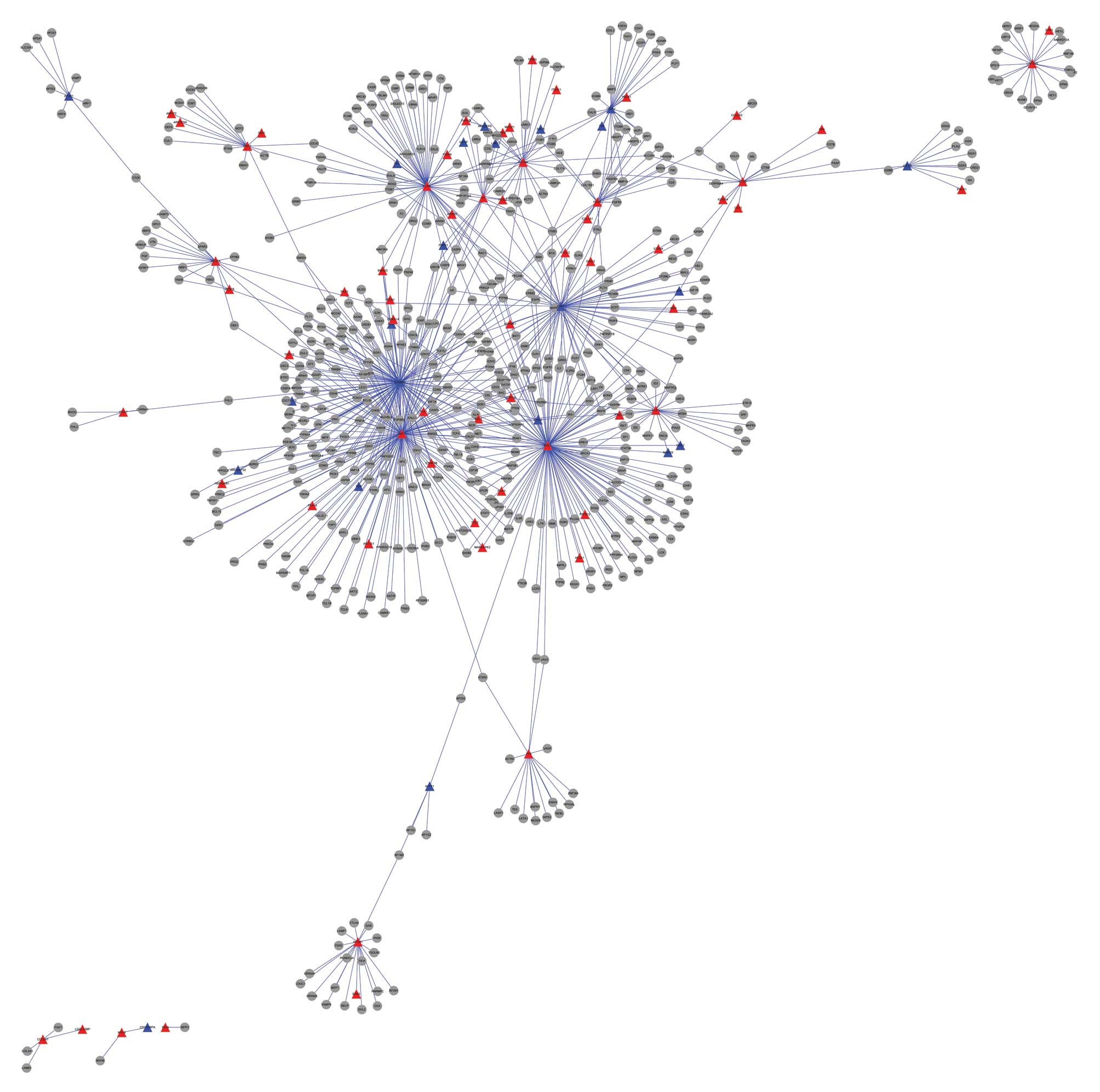
Pietschmann P, Rauner M, Sipos W and
Kerschan-Schindl K: Osteoporosis: an age-related and
gender-specific disease - a mini-review. Gerontology. 55:3–12.
2009. View Article : Google Scholar
|
|
5
|
Seeman E: Bone quality: the material and
structural basis of bone strength. J Bone Miner Metab. 26:1–8.
2008. View Article : Google Scholar
|
|
6
|
Kong YY, Yoshida H, Sarosi I, et al: OPGL
is a key regulator of osteoclastogenesis, lymphocyte development
and lymph-node organogenesis. Nature. 397:315–323. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sims AM, Shephard N, Carter K, et al:
Genetic analyses in a sample of individuals with high or low BMD
shows association with multiple Wnt pathway genes. J Bone Miner
Res. 23:499–506. 2008. View Article : Google Scholar
|
|
8
|
Kramer I, Keller H, Leupin O and Kneissel
M: Does osteocytic SOST suppression mediate PTH bone anabolism?
Trends Endocrinol Metab. 21:237–244. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Rawadi G, Vayssière B, Dunn F, Baron R and
Roman-Roman S: BMP-2 controls alkaline phosphatase expression and
osteoblast mineralization by a Wnt autocrine loop. J Bone Miner
Res. 18:1842–1853. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Almeida M, Han L, Bellido T, Manolagas SC
and Kousteni S: Wnt proteins prevent apoptosis of both uncommitted
osteoblast progenitors and differentiated osteoblasts by
beta-catenin-dependent and -independent signaling cascades
involving Src/ERK and phosphatidylinositol 3-kinase/AKT. J Biol
Chem. 280:41342–41351. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Jin H, Stewart TL, Hof RV, Reid DM, Aspden
RM and Ralston S: A rare haplotype in the upstream regulatory
region of COL1A1 is associated with reduced bone quality and hip
fracture. J Bone Miner Res. 24:448–454. 2009. View Article : Google Scholar
|
|
12
|
Kilian KA, Bugarija B, Lahn BT and Mrksich
M: Geometric cues for directing the differentiation of mesenchymal
stem cells. Proc Natl Acad Sci USA. 107:4872–4877. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Valtieri M and Sorrentino A: The
mesenchymal stromal cell contribution to homeostasis. J Cell
Physiol. 217:296–300. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Griffin M, Iqbal SA and Bayat A: Exploring
the application of mesenchymal stem cells in bone repair and
regeneration. J Bone Joint Surg Br. 93:427–434. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Jones E and Yang X: Mesenchymal stem cells
and bone regeneration: current status. Injury. 42:562–568. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Benisch P, Schilling T, Klein-Hitpass L,
et al: The transcriptional profile of mesenchymal stem cell
populations in primary osteoporosis is distinct and shows
overexpression of osteogenic inhibitors. PLoS One. 7:e451422012.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Irizarry RA, Bolstad BM, Collin F, Cope
LM, Hobbs B and Speed TP: Summaries of affymetrix GeneChip probe
level data. Nucleic Acids Res. 31:e152003. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Benjamini Y and Hochberg Y: Controlling
the false discovery rate: a practical and powerful approach to
multiple testing. J Roy Statist Soc Ser B Stat (Methodological).
57:289–300. 1995.
|
|
19
|
Ashburner M, et al: Gene Ontology: tool
for the unification of biology. Nature genetics. 2000.25(1): 25–29.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
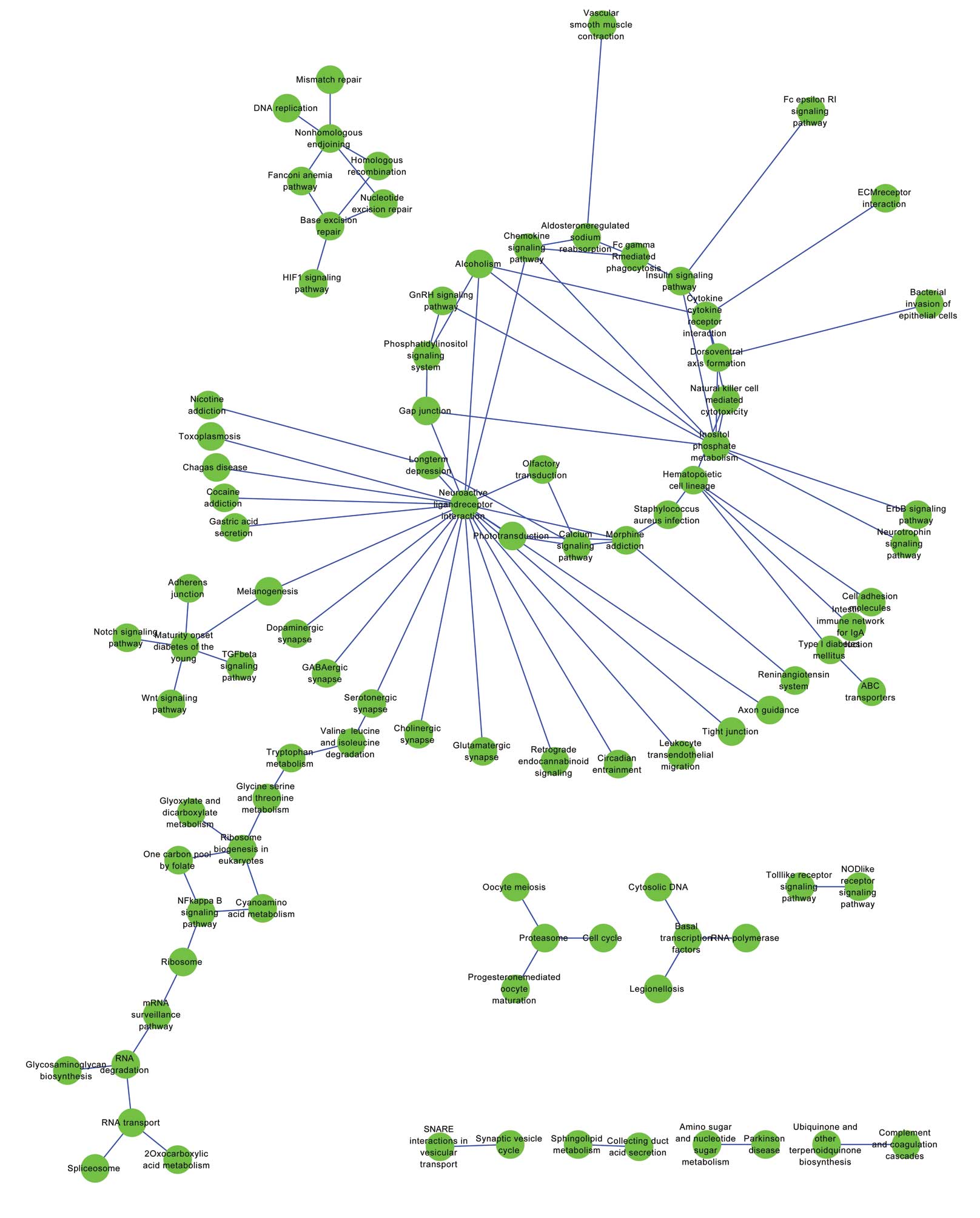
Kanehisa M and Goto S: KEGG: kyoto
encyclopedia of genes and genomes. Nucleic acids research.
2000.28(1): 27–30. View Article : Google Scholar
|
|
21
|
Da Wei Huang BTS and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nature protocols. 2008.4(1): 44–57.
View Article : Google Scholar
|
|
22
|
Hosack DA, Dennis G Jr, Sherman BT, Lane
HC and Lempicki RA: Identifying biological themes within lists of
genes with EASE. Genome Biol. 4:R702003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Stelzl U, Worm U, Lalowski M, et al: A
human protein-protein interaction network: a resource for
annotating the proteome. Cell. 122:957–968. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Przulj N, Wigle DA and Jurisica I:
Functional topology in a network of protein interactions.
Bioinformatics. 20:340–348. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wingender E, Dietze P, Karas H and Knüppel
R: TRANSFAC: a database on transcription factors and their DNA
binding sites. Nucleic Acids Res. 24:238–241. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li WF, Hou SX, Yu B, Li MM, Férec C and
Chen JM: Genetics of osteoporosis: accelerating pace in gene
identification and validation. Hum Genet. 127:249–285. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Jin H and Ralston SH: Regulatory
polymorphisms and osteoporosis. Gene Regulatory Sequences and Human
Disease. Springer; New York, NY: pp. p41–p54. 2012
|
|
28
|
Xu B, Song G and Ju Y: Effect of focal
adhesion kinase on the regulation of realignment and tenogenic
differentiation of human mesenchymal stem cells by mechanical
stretch. Connect Tissue Res. 52:373–379. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zintzaras E, Doxani C, Koufakis T,
Kastanis A, Rodopoulou P and Karachalios T: Synopsis and
meta-analysis of genetic association studies in osteoporosis for
the focal adhesion family genes: the CUMAGAS-OSTEOporosis
information system. BMC Med. 9:92011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Westendorf JJ, Kahler RA and Schroeder TM:
Wnt signaling in osteoblasts and bone diseases. Gene. 341:19–39.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Lau KH, Kapur S, Kesavan C and Baylink DJ:
Up-regulation of the Wnt, estrogen receptor, insulin-like growth
factor-I, and bone morphogenetic protein pathways in C57BL/6 J
osteoblasts as opposed to C3H/HeJ osteoblasts in part contributes
to the differential anabolic response to fluid shear. J Biol Chem.
281:9576–9588. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Otero K, Shinohara M, Zhao H, et al: TREM2
and β-catenin regulate bone homeostasis by controlling the rate of
osteoclasto-genesis. J Immunol. 188:2612–2621. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Rangaswami H, Schwappacher R, Tran T, et
al: Protein kinase G and focal adhesion kinase converge on
Src/Akt/β-catenin signaling module in osteoblast
mechanotransduction. J Biol Chem. 287:21509–21519. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Altintaş A, Saruhan-Direskeneli G, Benbir
G, Demir M and Purisa S: The role of osteopontin: a shared pathway
in the pathogenesis of multiple sclerosis and osteoporosis? J
Neurol Sci. 276:41–44. 2009. View Article : Google Scholar
|
|
35
|
Robertson Remen KM, Gustafsson JÅ and
Andersson G: The liver X receptor promotes macrophage
differentiation and suppresses osteoclast formation in mouse
RAW264.7 promy-elocytic leukemia cells exposed to bacterial
lipopolysaccharide. Biochem Biophys Res Commun. 430:375–380. 2013.
View Article : Google Scholar
|
|
36
|
Grant SF, Reid DM, Blake G, Herd R,
Fogelman I and Ralston SH: Reduced bone density and osteoporosis
associated with a polymorphic Sp1 binding site in the collagen type
I alpha 1 gene. Nat Genet. 14:203–205. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Jochum W, David JP, Elliott C, et al:
Increased bone formation and osteosclerosis in mice overexpressing
the transcription factor Fra-1. Nat Med. 6:980–984. 2000.
View Article : Google Scholar : PubMed/NCBI
|