Introduction
Congenital infection with human cytomegalovirus
(HCMV), which belongs to the herpesviridae group, and may be termed
human herpes virus-5, is the most common intrauterine infection
(1,2). Studies have demonstrated an
association between active CMV infection of the mother and in
utero HCMV transmission. The risk of congenital HCMV infection
is higher in infants when the mother acquires an initial CMV
infection during pregnancy, compared with that in infants when the
mother acquires the infection prior to conception (3–7).
Therefore, maternal antibodies against HCMV provide protection
against congenital infection (8,9).
Congenital HCMV infection poses a high risk of causing congenital
disorders. Congenital HCMV infection (15–20%) leads to long-term
disability including sensorineural hearing loss, visual impairment,
mental retardation and cognitive defects. Furthermore, 4% of
CMV-infected infants do not survive (3–5,8,10).
Understanding the association between congenital
infection and active maternal HCMV infection during pregnancy is
important for maternal and neonatal healthcare. Therefore, the
identification of active HCMV infection during pregnancy is
required. However, >95% pregnant females with primary CMV
infection are asymptomatic and, therefore, clinical diagnosis is
challenging (11,12). Seroconversion may be used to detect
HCMV antibodies during pregnancy. However, it is rarely effective,
due to the lack of antibody screening prior to conception, which
would enable the identification of seronegativity. Routine viral
culturing may be sufficiently sensitive for the identification of
CMV. However, this method is labor-intensive and subjective, and it
may take >14 days for the virus to be propagated and identified
(13). Due to the limitations of
culture-based methods, targeting the viral genome via quantitative
polymerase chain reaction (qPCR) has become an important laboratory
tool for the diagnosis and treatment of CMV infection. Previous
studies have evaluated qPCR for the detection and quantification of
CMV in plasma samples (14,15).
A novel nucleic acid amplification method,
loop-mediated isothermal amplification (LAMP), has been reported to
detect CMV viral genomic DNA (16). This method has been used for the
rapid diagnosis of a number of infectious diseases, including
herpes viruses (17–19), Epstein-Barr virus (20), hepatitis B Virus (21) and CMV (22). LAMP is capable of amplifying
specific sequences of DNA under homoeothermic conditions and
requires relatively simple and cost-effective equipment, making it
amenable for use in hospital laboratories.
In the present study, a simple LAMP assay was
established for the detection of CMV in peripheral blood samples
from pregnant women. This detection method exhibits the potential
for use in point-of-care settings for CMV infection screening and
follow-up during pregnancy.
Materials and methods
Clinical specimens and DNA
extraction
Whole blood samples from 336 pregnant women, who
were registered at Yantai Yuhuangding Hospital of Qingdao
University (Yantai, China), were used in the present study.
Amniotic fluid samples (11) were
obtained from pregnant women exhibiting CMV-positivity, which was
identified using reverse transcription qPCR (RT-qPCR) and LAMP
assays. Informed consent was obtained from the patients, and the
study was permitted by the Human Research Ethics Committee of
Yantai Yuhuangding Hospital of Qingdao University. Samples were
initially detected using RT-qPCR and then evaluated using a LAMP
assay. Total DNA from whole blood samples or amniotic fluid samples
was extracted using a QIAamp DNA Mini kit (Qiagen GmbH, Hilden,
Germany), according to the manufacturer’s instructions. Total
extracted DNA was quantified by measurements at 260 nm optical
density (OD) using a spectrophotometer (NanoDrop 2000; Thermo
Fisher Scientific, Wilmington, DE, USA). Extracted DNA samples were
stored at −20°C prior to use. Viral DNA isolation was performed
from stock viruses of herpes simplex virus type 1 (HSV-1), HSV-2,
varicella zoster virus (VZV), HSV-6, HSV-7 and CMV (Sinobio,
Beijing, China) using the QIAamp DNA Mini kit.
Primer design for LAMP
The primers for LAMP amplification of the CMV
glycoprotein B (gB) gene were designed based on CMV sequence data
obtained from Genbank (accession number: M60931). Oligonucleotide
primers that were used in the present study were designed using
Primer Explorer V4 software (Eiken Chemical Co. Ltd., Tokyo,
Japan). Designed primer sequences were subjected to BLAST
(http://blast.ncbi.nlm.nih.gov/Blast.cgi) in order to
exclude the possibility of cross-reactivity with HSV-1, HSV-2, VZV,
HSV-6 and HSV-7. CMV specific primers consisted of two outer (F3
and B3) and two inner primers: Forward inner primer (FIP) and
backward inner primer (BIP). Inner primers that recognized both
forward and reverse strands of the target DNA were connected by a
‘TTTT’ linker. And additional loop primers [forward loop primer
(LF) and backward loop primer (LB)] were used to promote both the
amplification efficiency and acceleration of the reaction. Details
of the sequence and location of each nucleotide primer in the
target DNA sequences are provided in Fig. 1.
 | Figure 1Locations and target sequences of the
CMV gB gene and the primers for CMV LAMP. (A) Target sequences in
the CMV gB gene. (B) Primer sequences for the CMV LAMP. F3, labeled
sequence in the target sequence; B3, reverse complementary sequence
in the target sequence; FIB, forward internal primer, reverse
complementary sequence of F1 + TTTT + labeled F2 sequence; BIP,
backwarrd internal primer, labeled B1 sequence + TTTT + reverse
complementary sequence of B2; LF, forward loop primer, labeled LPF
sequence in the target sequence; LB, backward loop primer, reverse
complementary sequence of labeled LPB sequence; CMV,
cytomegalovirus; LAMP, loop-mediated isothermal amplification;
glycoprotein B, gB. |
Optimization of LAMP conditions
In order to determine the sensitivity of the CMV
LAMP method, part of the gB gene containing the target DNA sequence
was amplified using the following primers: Forward
TGCCCGACGTCACGGTGGTC and reverse: ACCGACTTCAGGGTACTGG, which was
cloned into pGEM-T-Easy plasmid (Promega Corporation, Madison, WI,
USA). Optimization of LAMP conditions for CMV and sensitivity
determination was determined by amplifying
100–107 copies of CMV gB-containing plasmids.
The specificity of the LAMP assay was determined using HSV-1,
HSV-2, VZV, HSV-6 and HSV-7 DNA samples as negative controls.
Amplifications were optimized using different
conditions: Using 20, 25, 30 or 35 µl reaction volumes,
including 2, 5 or 10 µl DNA template, 1 or 2 µM inner
primers (FIP and BIP), 0.1, 0.3 or 0.5 µM outer primers (F3
and B3) and 0.5 or 1 µM loop primers (LF and LB), 0.5,1 or 2
µl Bst DNA polymerase (Large Fragment; New England Biolabs,
Inc., Ipswich, MA, USA), 2 × reaction mix (0.5 of the total
volume), and supplemented distilled and deionized water
(ddH2O). Reaction temperatures were screened at 59, 62
or 65°C and at the following reaction times: 5, 10, 15, 20, 25, 30
and 35 min. A LAMP turbidimeter TERAMECS (LA200; Teramecs, Co.
Ltd., Kyoto, Japan) was used to incubate the mixtures and to
measure the turbidity following the LAMP reaction. The turbidity
cut-off value was set at >0.1 mean ± 3 standard deviation of the
turbidity, from the turbidity values of three negative samples. The
LAMP products were also subjected to 1.5% agarose gel
electrophoresis in order to validate the experiments. Gels were
visualized under an ultraviolet light following ethidium bromide
staining.
CMV-specific RT-qPCR assay of whole-blood
and amniotic fluid samples
Primers for the CMV RT-qPCR assay were designed
according to previously reported sequences (23) and were synthesized by Sangon
Biotech (Shanghai, China). Primers were dissolved in
ddH2O, to 100 µM and stored at −20°C. The RT-qPCR
assay was performed using a One-Step PrimeScript RT-PCR kit (Takara
Bio, Inc., Otsu, Japan), using LightCycle 2.0 (Roche Diagnostics
GmbH, Mannheim, Germany).
Evaluation of LAMP with clinical
specimens
In order to evaluate the LAMP assay in whole blood
or amniotic fluid specimens from pregnant women, 336 whole blood
samples were tested for CMV using RT-qPCR. Whole blood samples
(336) and 11 amniotic fluid samples from RT-qPCR-confirmed
CMV-positive pregnant women were then subjected to a LAMP assay
using the optimized conditions (25 µl reaction volume,
including 3 µL DNA template, 1 µM inner primers, 0.5
µM outer primers and 0.5 µM loop primers, 1 µl
Bst DNA polymerase and ddH2O. The reaction was performed
at 62°C for 35 min). Sensitivity, specificity, positive predictive
value and negative predictive value from the LAMP assays were then
calculated using standard formulas and the results of RT-qPCR were
used as standards.
Results
Optimized conditions for LAMP assays
In order to optimize the conditions for CMV LAMP
detection, LAMP was conducted under different conditions, including
different Mg2+ concentrations, different concentrations
of loop primers, and different temperatures and durations. The
results suggested that LAMP conditions were optimized at a
25-µl reaction volume. Final reaction mixtures consisted of
1.6 µM inner primers (FIP and BIP), 0.2 µM outer
primers (F3 and B3), 0.8 µM loop primers (LF and LR), 10 mM
MgSO4, 1 µl Bst DNA polymerase and 5 µl
DNA template. Amplifications were performed at 64°C for 30 min and
reactions were terminated at 85°C for 5 min.
Specificity of LAMP using turbidity
assays and gel electrophoresis
In the present study, the capability of the CMV LAMP
assay for the discrimination of CMV from other members of
herpesviridae, such as HSV-1, HSV-2, VZV, HSV-6 and HSV-7 was
assessed. The results of the present study suggested that CMV
exhibits bright turbidity following a LAMP assay, whereas other
members of herpesviridae were not detected using LAMP (Fig. 2A). LAMP products were then analyzed
using gel electrophoresis and the results suggested that CMV was
successfully amplified, while other viral DNA was not amplified
(Fig. 2B). Therefore, primer sets
developed in the present study exhibit specificities for the target
CMV sequences.
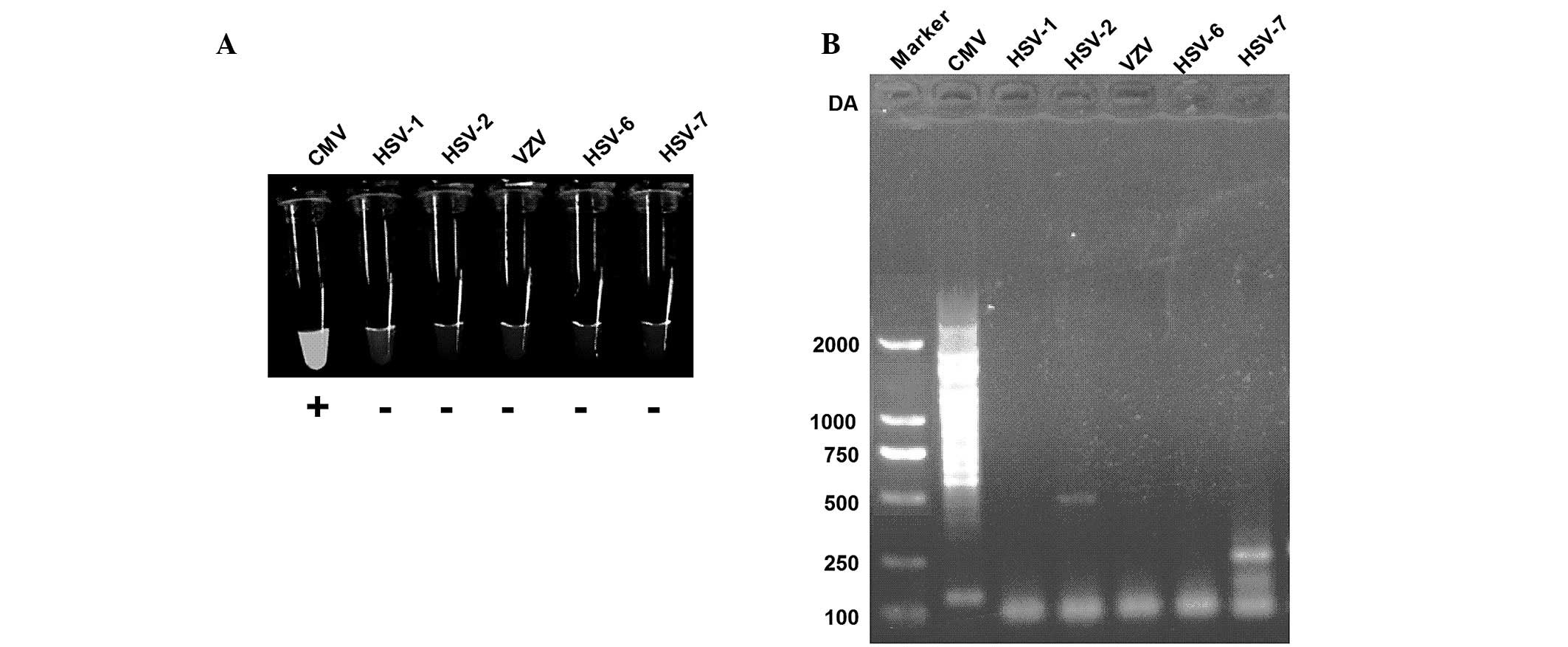 | Figure 2LAMP assay specificity to CMV,
targeted to the CMV gB gene. (A) Visual inspection of LAMP assay
for CMV, HSV-1, HSV-2, VZV, HSV-6 and HSV-7. (B) Electrophoretic
analysis of LAMP product from samples of CMV, HSV-1, HSV-2, VZV,
HSV-6 and HSV-7. LAMP, loop-mediated isothermal amplification; CMV,
cytomegalovirus; HSV, herpes simplex virus; VSV, varicella zoster
virus. |
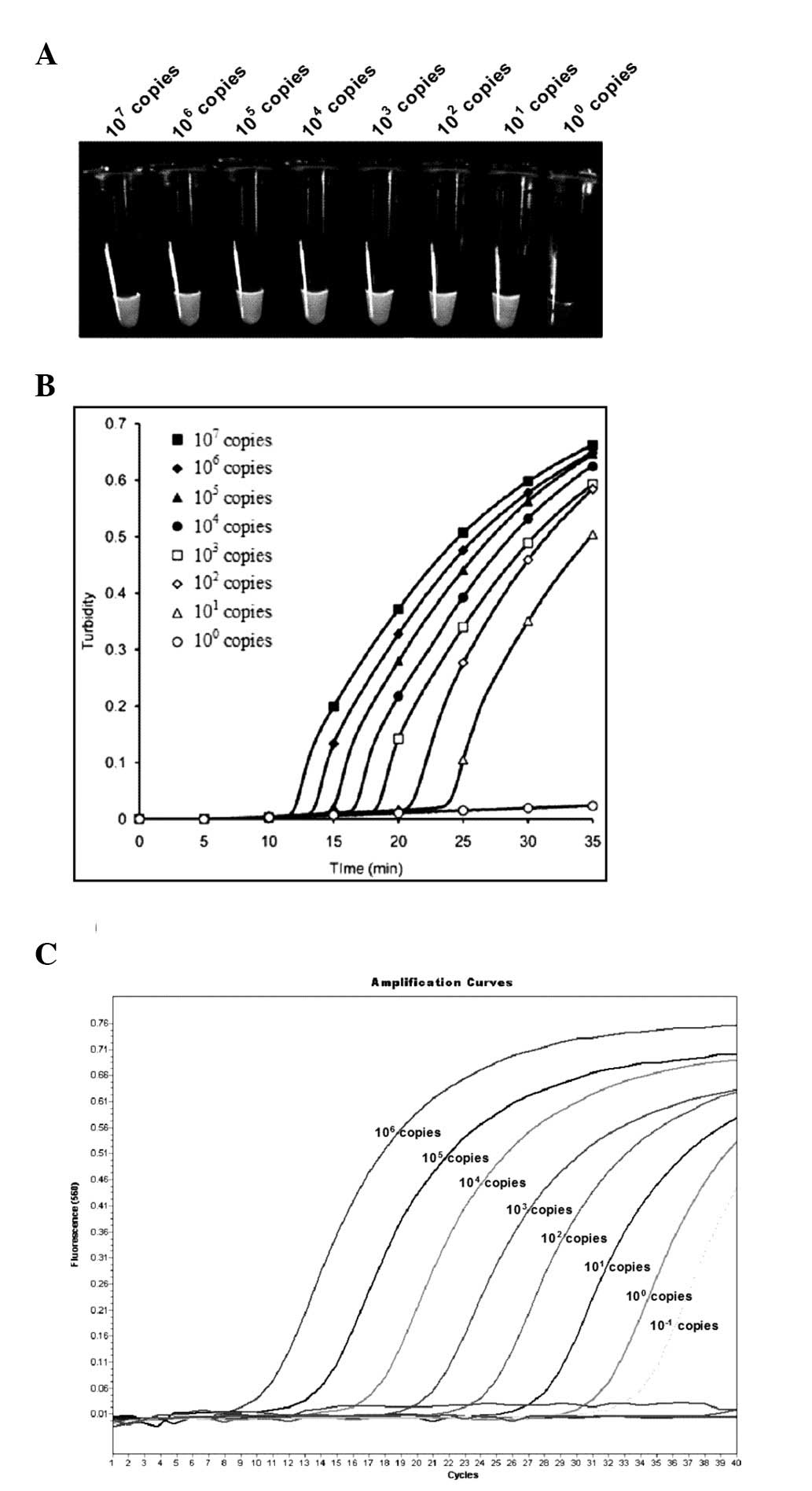
Sensitivity of LAMP using turbidity
measurement and RT-qPCR
LAMP assay sensitivity was analyzed using the
serially-diluted CMV gB-containing plasmid. Serial dilutions of
recombinant pGEM-T Easy plasmid ranging from
107–100 copies per tube were used in order to
determine the detection limits of CMV LAMP. Results demonstrated
that the sensitivity of the CMV LAMP assay was 10 copies per tube,
according to a real-time turbidimeter at 650 nm OD (Fig. 3A and B). Reactions were repeated
three times. Serially-diluted plasmids were examined, ranging from
106–10−1 copies per tube, using RT-qPCR
(Fig. 3C). The threshold of
RT-qPCR was 10−1 copies per tube, which was 10 times
more sensitive than that of the CMV LAMP assay.
CMV LAMP assay of whole blood specimens
from pregnant women (Tables I and
II)
In order to further evaluate the performance of the
CMV LAMP assay for CMV infection detection in pregnant women, CMV
LAMP assays were conducted in 336 whole blood samples and 11
amniotic fluid samples from pregnant women. Samples were tested
using RT-qPCR. Positive samples (10, 13, 20 or 21), with a
threshold of 10 copies, were confirmed using RT-qPCR.
RT-qPCR-positive samples, 10 and 13, were confirmed as positive
according to the CMV LAMP, with a threshold of 103 or
104 copies per tube, respectively and 100% sensitivity.
However, samples 2 and 3, which were negative according to RT-qPCR,
were shown to be positive according to the CMV LAMP assay, with a
specificity of 98.45 and 97.24%, PPV values of 86.67 and 76.92%,
respectively; NPV for both thresholds were 100% (Table I). The following values were
observed at 101 and 102 copies per tube
compared with those at 103 and 104:
Sensitivity decreased to 86.96 and 91.30%, specificity increased to
100 and 99.68%, PPV increased to 100 and 94.45%, and NPV decreased
to 99.05 or 99.36%, at 101 and 102 copies per
tube respectively. In order to reconfirm the sensitivity and
specificity of the CMV LAMP assay, 11 amniotic fluid samples were
examined from pregnant women with whole blood CMV-positive and 15
control amniotic fluid samples were examined, from pregnant women
without active CMV infection. The results indicated that 11 samples
were positive, with a threshold of 102, 103
or 104, while the 15 control samples were negative, with
a threshold of 101, 102 or 103
(Table II). Overall, the CMV LAMP
method performed well in the detection of CMV infection.
Discussion
LAMP reaction requires DNA polymerase with strand
displacement activity and >4 specifically designed primers.
During the first step, a stem-loop DNA structure is constructed, in
which the sequences of both DNA ends are derived from the inner
primers. Subsequently, one inner primer hybridizes to the loop on
the LAMP cycle product and initiates strand displacement DNA
synthesis, yielding the original stem-loop and new stem-loop DNA
with a stem that is twice as long. The final products are termed
stem-loop DNAs, and have several inverted repeats of the target DNA
and cauliflower-like structures with multiple loops, amplifying
<109 copies of the target. LAMP is a rapid and simple
technique for the amplification of specific DNA sequences that has
advantages over PCR (24,25). The most significant advantage of
LAMP is its ability to amplify specific sequences of DNA at a
constant temperature (63–65°C), without thermocycling. In addition,
<45 min are required in order to amplify the target sequences.
Given these advantages, LAMP may be adopted for widespread use in
hospital laboratories.
In the present study a CMV LAMP assay was
established for the detection of CMV DNA in pregnant women with CMV
infection, using RT-qPCR in order to confirm active CMV infection.
Following optimization of the PCR protocol, the components for CMV
LAMP included a 25-µl reaction volume with 1.6 µM
inner primers, 0.2 µM outer primers, 0.8 µM loop
primers, 10 mM MgSO4, 1 µl Bst DNA polymerase and
5 µl DNA template. The amplification was conducted at 64°C
for 30 min. This PCR method was specific for the amplification of
CMV DNA and it did not amplify HSV-1, HSV-2, VZV, HSV-6 or HSV-7,
which belong to the same herpesviridae family. Sensitivity
determination using the serially-diluted CMV gB-containing plasmids
demonstrated that >10 copies per tube were detectable using the
CMV LAMP method, which had a 10 fold lower sensitivity level
compared with that of RT-qPCR.
Furthermore, the CMV LAMP assay performed well in
the detection of CMV infection. The detection results for 336 whole
blood samples demonstrated that, at a threshold of
101–104 copies per tube, the sensitivity of
the LAMP assay for the detection of CMV infection was 86.96–100%,
specificity was 97.24–100%, PPV was 76.92–100% and NPV was
99.05–100%. The LAMP assay was sensitive and specific for the
detection of CMV in 11 amniotic fluid samples from CMV-positive
pregnant women and in 15 control amniotic fluid samples. Overall,
the CMV LAMP method performed well in the detection of CMV
infection.
In conclusion, a CMV LAMP method was developed,
which was highly sensitive, specific, simple and timesaving.
Furthermore, it performed well in the detection of active CMV
infection in pregnant women. Therefore, the present study provides
novel insights into the detection of active CMV infection in
pregnant women.
Acknowledgments
The present study was supported by a grant from
Yantai Yuhuangding Hospital of Qingdao University.
References
|
1
|
Adler SP and Marshall B: Cytomegalovirus
infections. Pediatr Rev. 28:92–100. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Enders G, Daiminger A, Bäder U, Exler S
and Enders M: Intrauterine transmission and clinical outcome of 248
pregnancies with primary cytomegalovirus infection in relation to
gestational age. J Clin Virol. 52:244–246. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Coll O, Benoist G, Ville Y, Weisman LE,
Botet F, Anceschi MM, Greenough A, Gibbs RS and Carbonell-Estrany
X; WAPM Perinatal Infections Working Group: Guidelines on CMV
congenital infection. J Perinat Med. 37:433–445. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Revello MG, Fabbri E, Furione M, Zavattoni
M, Lilleri D, Tassis B, Quarenghi A, Cena C, Arossa A, Montanari L,
et al: Role of prenatal diagnosis and counseling in the management
of 735 pregnancies complicated by primary human cytomegalovirus
infection: A 20-year experience. J Clin Virol. 50:303–307. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Guerra B, Simonazzi G, Banfi A, Lazzarotto
T, Farina A, Lanari M and Rizzo N: Impact of diagnostic and
confirmatory tests and prenatal counseling on the rate of pregnancy
termination among women with positive cytomegalovirus
immunoglobulin M antibody titers. Am J Obstet Gynecol. 196:e1–e6.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Fowler KB, Stagno S, Pass RF, Britt WJ,
Boll TJ and Alford CA: The outcome of congenital cytomegalovirus
infection in relation to maternal antibody status. N Engl J Med.
326:663–667. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Stagno S, Pass RF, Cloud G, Britt WJ,
Henderson RE, Walton PD, Veren DA, Page F and Alford CA: Primary
cytomegalovirus infection in pregnancy. Incidence, transmission to
fetus, and clinical outcome. JAMA. 256:1904–1908. 1986. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Manicklal S, Emery VC, Lazzarotto T,
Boppana SB and Gupta RK: The ‘silent’ global burden of congenital
cytomegalovirus. Clin Microbiol Rev. 26:86–102. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Revello MG and Gerna G: Pathogenesis and
prenatal diagnosis of human cytomegalovirus infection. J Clin
Virol. 29:71–83. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kenneson A and Cannon MJ: Review and
meta-analysis of the epidemiology of congenital cytomegalovirus
(CMV) infection. Rev Med Virol. 17:253–276. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Revello MG and Gerna G: Diagnosis and
management of human cytomegalovirus infection in the mother, fetus,
and newborn infant. Clin Microbiol Rev. 15:680–715. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lazzarotto T, Guerra B, Lanari M,
Gabrielli L and Landini MP: New advances in the diagnosis of
congenital cytomegalovirus infection. J Clin Virol. 41:192–197.
2008. View Article : Google Scholar
|
|
13
|
de Vries JJ, van der Eijk AA, Wolthers KC,
Rusman LG, Pas SD, Molenkamp R, Claas EC, Kroes AC and Vossen AC:
Real-time PCR versus viral culture on urine as a gold standard in
the diagnosis of congenital cytomegalovirus infection. J Clin
Virol. 53:167–170. 2012. View Article : Google Scholar
|
|
14
|
Boaretti M, Sorrentino A, Zantedeschi C,
et al: Quantification of cytomegalovirus DNA by a fully automated
real-time PCR for early diagnosis and monitoring of active viral
infection in solid organ transplant recipients. J Clin Virol.
56:124–128. 2013. View Article : Google Scholar
|
|
15
|
Bravo D, Clari MÁ, Costa E, Muñoz-Cobo B,
Solano C, José Remigia M and Navarro D: Comparative evaluation of
three automated systems for DNA extraction in conjunction with
three commercially available real-time PCR assays for quantitation
of plasma cytomegalovirus DNAemia in allogeneic stem cell
transplant recipients. J Clin Microbiol. 49:2899–2904. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Notomi T, Okayama H, Masubuchi H, Yonekawa
T, Watanabe K, Amino N and Hase T: Loop-mediated isothermal
amplification of DNA. Nucleic Acids Res. 28:E632000. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Enosawa M, Kageyama S, Sawai K, et al: Use
of loop-mediated isothermal amplification of the IS900 sequence for
rapid detection of cultured Mycobacterium avium subsp
paratuberculosis. J Clin Microbiol. 41:4359–4365. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Iwamoto T, Sonobe T and Hayashi K:
Loop-mediated isothermal amplification for direct detection of
Mycobacterium tuberculosis complex, M. avium, and M. intracellulare
in sputum samples. J Clin Microbiol. 41:2616–2622. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hagiwara M, Sasaki H, Matsuo K, Honda M,
Kawase M and Nakagawa H: Loop-mediated isothermal amplification
method for detection of human papillomavirus type 6, 11, 16, and
18. J Med Virol. 79:605–615. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Iwata S, Shibata Y, Kawada J, Hara S,
Nishiyama Y, Morishima T, Ihira M, Yoshikawa T, Asano Y and Kimura
H: Rapid detection of Epstein-Barr virus DNA by loop-mediated
isothermal amplification method. J Clin Virol. 37:128–133. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Nyan DC, Ulitzky LE, Cehan N, Williamson
P, Winkelman V, Rios M and Taylor DR: Rapid detection of hepatitis
B virus in blood plasma by a specific and sensitive loop-mediated
isothermal amplification assay. Clin Infect Dis. 59:16–23. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Suzuki R, Yoshikawa T, Ihira M, Enomoto Y,
Inagaki S, Matsumoto K, Kato K, Kudo K, Kojima S and Asano Y:
Development of the loop-mediated isothermal amplification method
for rapid detection of cytomegalovirus DNA. J Virol Methods.
132:216–221. 2006. View Article : Google Scholar
|
|
23
|
Nefzi F, Ben SN, Khelif A, Feki S, Aouni M
and Gautheret-Dejean A: Quantitative analysis of human
herpesvirus-6 and human cytomegalovirus in blood and saliva from
patients with acute leukemia. J Med Virol. 87:451–460. 2015.
View Article : Google Scholar
|
|
24
|
Notomi T, Okayama H, Masubuchi H, Yonekawa
T, Watanabe K, Amino N and Hase T: Loop-mediated isothermal
amplification of DNA. Nucleic Acids Res. 28:E632000. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Nagamine K, Hase T and Notomi T:
Accelerated reaction by loop-mediated isothermal amplification
using loop primers. Mol Cell Probes. 16:223–229. 2002. View Article : Google Scholar : PubMed/NCBI
|