Introduction
Alternative splicing (AS) increases the complexity
of eukaryotic gene expression and causes the diversification of
protein function. Furthermore, 92–94% of human multi-exon genes
undergo alternative splicing (1,2).
Fragile X mental retardation 1 gene (FMR1) is associated
with fragile X syndrome (FXS). The gene is located on chromosome
Xq27.3, is 38 kb long, and consists of 17 exons and 16 introns.
FMR1 AS gene expression has been observed in a number of
different human and mouse tissues (3–6).
Previous research has suggested that AS of the FMR1 gene
involves the inclusion or exclusion of exons 12 and 14, and the
selection of 3′ splice sites in exons 15 and 17, which may generate
up to 24 different mature transcripts. To date, 20 spliced isoforms
(ISO; 1–20) have been reported (3,4,6,7).
Spliced isoforms that are abundantly expressed are
more likely to affect gene activity and patient physiology,
compared with those that are expressed in low abundance (8). Analysis of the primary splice
variant(s) that are expressed in different tissues and cell types
is required, in order to understand the functions of alternatively
spliced genes. Using reverse transcription-polymerase chain
reaction (RT-PCR) and western blotting, a number of different
alternatively spliced products have been observed in FMR1
gene expression, in numerous types of tissue and cell lines.
However, to the best of our knowledge, the proportion of spliced
isoforms, expressed in different types of tissues and cells
(3–7), has not been investigated. ISO7
(lacking exon 12) was found to be the predominant isoform expressed
in COS cells and in lymphoblasts. By contrast, ISO1 (containing all
exons) was not detected in any cell types, suggesting that ISO1
expression is very rare or absent in COS cells (7). Studies have demonstrated that ISO13
(containing the first splicing acceptor site in exon 17) is the
dominant isoform in human heart, spleen, liver, kidney and fetal
cerebral cortex tissues. ISO13 accounted for approximately 40% of
all spliced isoforms in human fetal brain neurons, whereas other
isoforms, including ISO1, were relatively rare (9,10).
ISO8 (lacking exon 12 and containing the second splicing acceptor
site in exon 15) is dominant in human adult cerebral cortex tissue,
in contrast with that of human fetal cerebral cortex tissue
(10). These data suggest that the
predominant splice variants vary between different types of tissues
and cells. Therefore, analyses of FMR1 gene AS profiles are
required in order to further understand FXS pathogenesis, in
particular in FXS-affected tissues, such as the brain and
testis.
The FMR1 gene AS site is located at ~700 bp.
The difference in size between alternatively spliced products is
small. It is, therefore, difficult to distinguished AS sites using
routine RT-PCR or northern blotting (10). A combined method, using RT-PCR and
cloning, has been developed in order to detect alternatively
spliced mRNA, and to analyze the splicing pattern of transcripts
and the proportion of different alternatively spliced products
(9,11,12).
In the present study, in order to analyze the AS expression of
FMR1 gene, a combined method, cloning and sequencing of the
entire coding sequence of the FMR1 gene by means of TA
cloning, was conducted in human peripheral blood, and brain and
testis tissues.
Materials and methods
RT-PCR of the FMR1 gene from human
peripheral blood
The present study was approved by the ethics
committee of Fuzhou General Hospital (Fuzhou, China). Peripheral
blood was collected from four healthy individuals following
attainment of written informed consent and total RNA was extracted
using an RNA isolation kit (Qiagen, Hilden, Germany). Total RNA
(800 ng) was reverse transcribed using reverse transcriptase
(Invitrogen, Life Technologies, Carlsbad, CA, USA). A 2070 bp
fragment consisting of the entire coding region of the FMR1
cDNA was amplified using the following primers: Forward:
5′-TCGAGCGCCCGCAGCCCACCTCT-3′, and reverse:
5′-TGCCCTGTGCCATCTTGCCTACTATTT-3′. The PCR mixture (50 µl)
consisted of cDNA (2 µl) and Taq DNA polymerase (5 U). PCR
conditions were as follows: Initial denaturation at 94°C for 5 min,
30 cycles of denaturation at 94°C for 30 sec, annealing at 72°C for
3 min and extension at 72°C for 7 min. All RT-PCR products were
electrophoresed on 1% agarose gels (Sangon Biotech, Shanghai,
China) and extracted using a QIAquick gel extraction kit
(Qiagen).
RT-PCR of the FMR1 gene from human brain
and testis cDNA
Human brain and testis QUICK-Clone cDNA was
purchased from Clontech Laboratories, Inc. (Mountainview, CA, USA).
Human whole brain and testis RNA was pooled from 8 and 39 healthy
male Caucasians, respectively, who had died unexpectedly. The cDNA
was used in order to amplify the entire coding region of the
FMR1 gene, as described for the human peripheral blood.
T cloning-sequencing of RT-PCR
products
Purified long chain RT-PCR products were ligated to
pGEM-T Easy vector using an pGEM-T Easy vector system (Promega
Corporation, Madison, WI, USA), according to the manufacturer’s
instructions. Subsequently, XL1-blue cells were transformed and the
recombinants were selected on Luria-Bertani
broth/ampicillin/isopropyl β-D-1-thiogalactop yranoside/X-Gal
plates (all Sangon Biotech). Recombinants were identified using
restrictive digestion with EcoRI and sent to a
Shanghai-based company (Sangon Biotech), in order to sequence the
inserts with Sanger sequencing.
Evolutionary selection pressure analysis
of the FMR1 exons
Vertebrate genome sequences (total 46, including
human) were used for selection pressure analysis of the FMR1
gene exons. Genome alignment information used in the present study
was obtained from the UCSC (http://genome.ucsc.edu/). Vertebrate genome alignments
(total 45) with human for coding DNA sequence (CDS) regions were
obtained from ftp://hgdownload.cse.ucsc.edu/ (13). The aligned exon sequences (45
vertebrate genome alignments with human for CDS regions) were
downloaded from the UCSC genome browser. Aligned exon sequences
were then manually corrected according to the protein alignment
(exon lengths were in multiples of three) and multiple sequence
alignment analysis was then performed using MEGA 5.0 (14). Subsequently, the ratio of
nonsynonymous substitutions per nonsynonymous site (dN) to
synonymous substitutions per synonymous site (dS; dN/dS) was
estimated using the on-line DataMonkey package with
likelihood-based methods (http://www.datamonkey.org), in order to measure
selection pressure on amino acid sequences (15). A conservative single-likelihood
ancestor-counting (SLAC) method was then conducted (16).
Results
Profiles of alternatively spliced
products of the FMR1 gene in human tissues
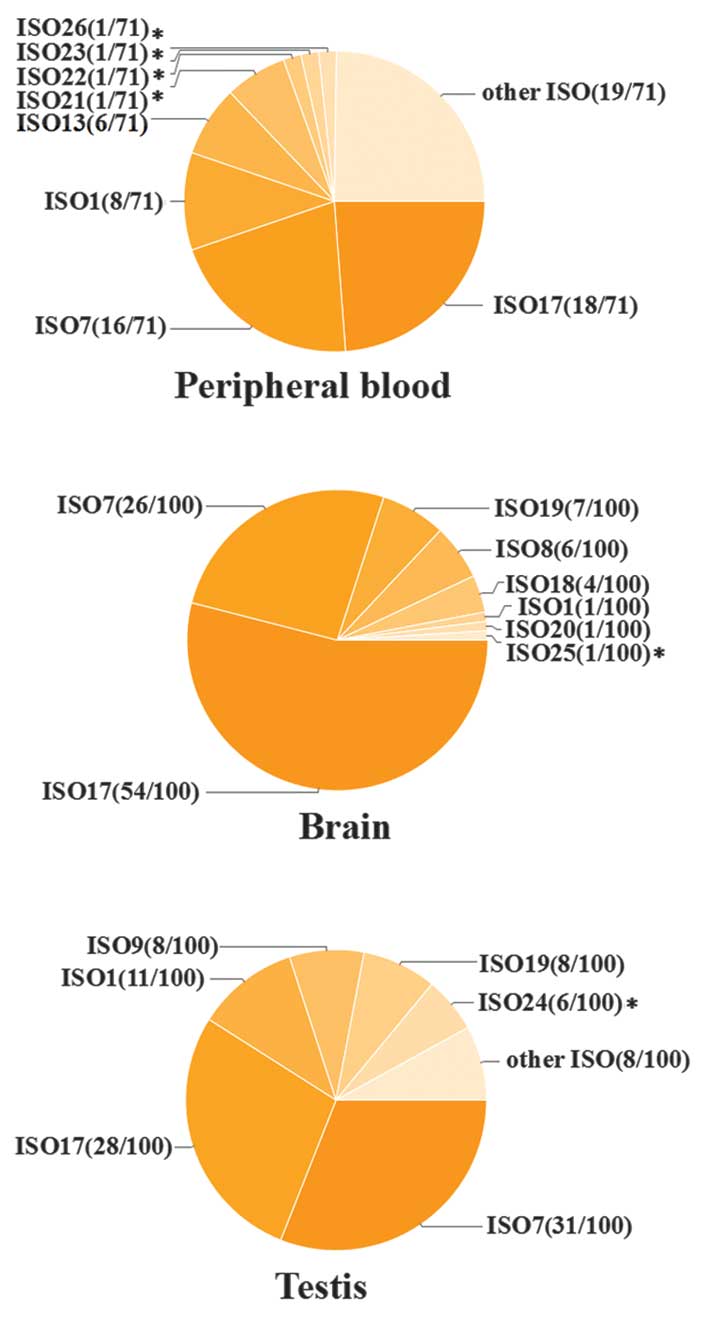
Recombinant products were randomly selected, with
71, 100 and 100 from RT-PCR products of human peripheral blood,
brain and testis, respectively. Sequencing results indicated that
20 different spliced products of the FMR1 gene were
identified among the 271 recombinants (Fig. 1); 20 isoforms were identified that
were not entirely consistent with those previously reported
(3–6). Nomenclature in Fig. 1 is derived from that of Sittler
et al (7). Six spliced
products, ISO2, ISO4, ISO5, ISO6, ISO11 and ISO16, were not
observed in the present study, suggesting that these six isoforms
may be expressed in low levels in human peripheral blood, and brain
and testis tissues.
ISO21–23 and ISO26 were only identified in
peripheral blood sequences, whereas ISO24 and ISO25 were only
observed in testis and brain sequences. ISO21 and ISO22 adopt
different combinations to those splicing patterns in previous
studies (7,9). Furthermore, they may be included in
the predicted 24 products (Fig.
1). ISO21, skipping exon 14, with the first splicing acceptor
site in exon 15 and the second splicing acceptor site in selected
exon 17, may encode a truncated polypeptide consisting of 439 amino
acid residues with a different carboxyl terminus, as compared with
the reference protein sequence of human FMR1 (GenBank ID:
AAB28395.2). ISO22, skipping exon 12, may encode a truncated
polypeptide of 428 amino acid residues with a different carboxyl
terminus. ISO23–26 were the result of the four novel splicing
sites, whose sequences have been deposited in the GenBank database.
ISO23 (GenBank ID: KC774029), skipping exons 11 and 12, with the
second splicing acceptor site of selected exon 17, may encode a
truncated polypeptide of 544 amino acid residues with a different
carboxyl terminus. ISO24 (GenBank ID: KF856234), with the second
splicing acceptor site of selected exon 15, may encode a truncated
polypeptide of 554 amino acid residues without changing the open
reading frame. ISO25 (GenBank ID: KC774027), with the novel
splicing acceptor site of exon 15 and the selected third downstream
site, may encode a polypeptide of 566 amino acid residues without
changing the open reading frame. ISO26 (GenBank ID: KC774028),
skipping exons 12 and 15, with a novel splicing donor site of exon
14 and a novel splicing acceptor site of selected exon 16, may
encode a truncated polypeptide of 418 amino acid residues with a
different carboxyl terminus (Fig.
2).
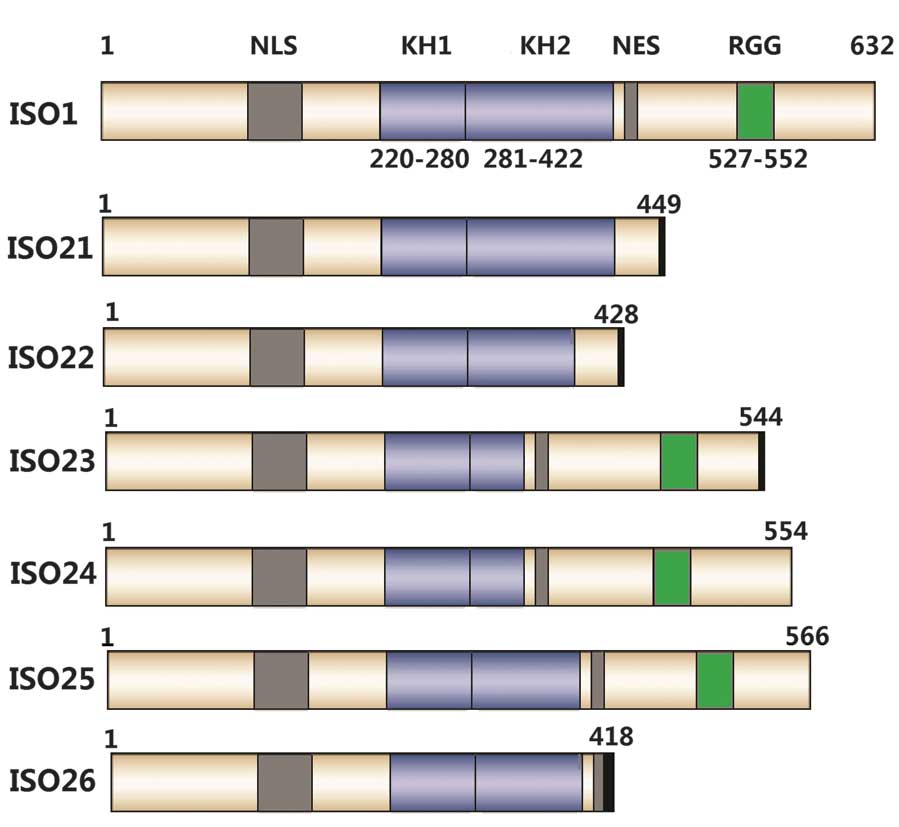 | Figure 2Predicted coding products of ISO1 and
six novel splicing transcripts (ISO21–ISO26). ISO1 represents the
FMR1 gene coding product with the full-length coding
sequence, comprising NLS, KH1, KH2, NES and RGG. Numbers represent
the location of amino acids. The black bar represents the carboxyl
terminus of the premature termination codon. ISO, isoform;
FMR1, fragile X mental retardation 1; NLS, nuclear
localization signal; NES, nuclear export signal; KH, K homology
domain; RGG, arginine-glycine-rich region. |
The results of the present study suggested that ~50%
of spliced isoforms are expressed at low levels, including those
that have previously been reported. For example, only one spliced
product of the following were observed: ISO9, 10 and 12, and the
novel isoforms, ISO21–23, 25 and 26.
Spliced products containing the full-length coding
sequence (ISO1) accounted for a low proportion of isoforms (7.38%;
20/271). In particular, in sequences derived from brain tissue
samples, ISO1 was detected once (1%; Fig. 3).
Alternatively spliced products lacking
exon 12 were predominant
The results of the analyses suggested that the
following isoforms were the most abundantly expressed: ISO7,
lacking exon 12, and ISO17, lacking exon 12, with the second
splicing acceptor site of exon 17 selected. However, the relative
abundance of the these spliced products varied between tissues. In
the brain, ISO17 was the predominant isoform (54%), whilst in the
testis, ISO7 was the predominant isoform (Fig. 3). In the brain, 99% of the isoforms
lacked exon 12, and ISO1, which consists of the entire coding
sequence was detected only once.
Exon 12 may be under strong positive
selection
According to the splicing pattern observed in the
human FMR1 gene, exons were classified into AS exons (exons
11–12, 14–17) and constitutive exons (exons 1–10,13). The results
of an SLAC analysis of 17 pre-aligned exon sequences suggested that
the mean dN/dS value for AS exons was 0.410, which was
significantly higher than that for the constitutive exons (0.067).
The results demonstrated that the dN/dS ratio of exon 12 was >1
(dN/dS=1.735), whilst that of all other exons was <1 (Table I).
 | Table IAnalysis of dN/dS ratios for 17 exons
in FMR1 gene. |
Table I
Analysis of dN/dS ratios for 17 exons
in FMR1 gene.
| Exon | dN/dS |
|---|
| Constitutive
exons | 0.067 |
| Exon 1 | 0.153 |
| Exon 2 | 0.058 |
| Exon 3 | 0.081 |
| Exon 4 | 0.031 |
| Exon 5 | 0.041 |
| Exon 6 | 0.112 |
| Exon 7 | 0.079 |
| Exon 8 | 0.018 |
| Exon 9 | 0.061 |
| Exon 10 | 0.066 |
| Exon 13 | 0.054 |
| Alternative
splicing exons | 0.410 |
| Exon 11 | 0.257 |
| Exon 12 | 1.735 |
| Exon 14 | 0.115 |
| Exon 15 | 0.092 |
| Exon 16 | 0.097 |
| Exon 17 | 0.165 |
Discussion
In the present study, 20 types of alternatively
spliced products were detected, six of which were previously
unreported (4–6,9,17,18).
Among these six alternatively spliced products, four novel splicing
patterns were identified, including an alternative 5′ splice site,
two alternative 3′ splice sites and an exon skipping site. In
addition, six previously reported spliced products were not
observed in the present study. These results indicate that the AS
of the FMR1 gene may be more complex than previously
understood.
In accordance with previous reports (7,9),
ISO1 was not the most abundant isoform observed in the peripheral
blood cells, and brain and testis tissues (7.38%; 20/271). Skipping
of exon 14 has been found to affect the subcellular localization of
the FMR protein (7). This suggests
that AS may be involved in the functioning and regulation of the
FMR1 gene. More abundant splice variants are more likely to
be associated with important gene functions, compared with those
which are less abundant (8). In
the present study, ISO7 and ISO17, lacking exon 12, were the
dominant splicing products. In the brain, these isoforms together
accounted for 80% of all isoforms (Fig. 3). Therefore, a lack of exon 12 may
be involved in the physiological functioning of the FMR1
gene.
FMR protein is an RNA-binding protein, abundant in
the brain and testis, which selectively binds to its own mRNA and
to ~4% of brain mRNA, which is associated with neurodevelopment and
synaptic plasticity (19,20). It has been demonstrated that the
discrepancy in FMR1 transcripts between fetal and adult
cerebral cortex tissue is associated with the presence of exon 12,
which is present in the fetal tissue but not in that of adults
(10). Exon 12 lengthens the
second K homology (KH) domain, an FMR1 RNA binding domain,
which may influence the specificity or affinity of FMR protein
RNA-binding (21). Exon 12
variants differentially interact with kissing complex RNA and
exhibit a distinct RNA-binding specificity (22,23).
Furthermore, it has been reported that the I304N mutation located
on the KH2 domain may lead to the expression of a serious FXS
phenotype (24). Studies have
shown that FMR1 with a I304N mutation exhibited a tenfold
average decrease in AUCK-containing RNA expression compared with
that in wild-type FMR1 (21). Therefore, AS products lacking exon
12 may shorten the second KH2 domain, which may influence the
specificity and affinity of FMR protein RNA-binding and eventually
target specific mRNA to regulate the corresponding protein
expression. The results of the present study suggested that AS
spliced products lacking exon 12 may be associated with the
physiological functioning of FMR1 gene.
Different types of selection pressure analyses have
been conducted in order to predict the function of AS, such as
dN/dS (or Ka/Ks) against amino acid mutations, and dS (or Ks)
against mutations at synonymous sites, with the former being the
most common (25,26). A genome-wide selection pressure
analysis in humans, chimpanzees, mice and rats has demonstrated
that Ka/Ks values were higher for AS exons compared with those for
constitutive exons, implying that AS events may accelerate protein
sequence evolution (27). In the
present study, dN/dS values for FMR1 exons demonstrated a
similar pattern. Furthermore, exon 12 exhibited markedly high (≥1)
dN/dS values compared with that for other exons. This is in
accordance with the results of a previous study that demonstrated
higher dN/dS values for CD45 gene AS exons compared with those for
constitutive exons, which suggests that the AS exons may be under
positive selection (28).
Therefore, exon 12, coding for part of the KH2 domain, may be under
strong positive selection and associated with vertebrate evolution.
Exon 12 consists of 63 nucleotides and, as this is a multiple of
three, it is a ‘modular’ exon that may be inserted or removed from
the transcripts without affecting the rest of the protein, which
may facilitate AS functioning (29).
ISO24 is a novel spliced product that was detected
in testis. It exhibited exon 11 skipping, which was also detected
in ISO23 (Fig. 1). Exon 11 encodes
part of the KH2 domain and consists of 135 nucleotides, and,
therefore, like exon 12, may represent a ‘modular’ exon. The dN/dS
ratio analysis suggested that exon 11 exhibited higher value in AS
exons, as compared with the other exons, except for exon 12
(Table I). The results of the
present study indicated that the skipping of exon 11 may represent
a functional AS event.
In conclusion, the results of the present study
implied that AS spliced products lacking exon 12 may be associated
with the physiological functioning the FMR1 gene. Further
investigation of the expression of isoforms ISO7 and ISO17 may help
to improve the understanding of the structure and function of the
FMR1 gene and the pathogenesis of FXS.
Acknowledgments
This study was supported by Key Scientific Projects
of Fujian Province, P.R. China (grant no. 2010Y0045).
References
|
1
|
Johnson JM, Castle J, Garrett-Engele P, et
al: Genome-wide survey of human alternative pre-mRNA splicing with
exon junction microarrays. Science. 302:2141–2144. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wang ET, Sandberg R, Luo S, et al:
Alternative isoform regulation in human tissue transcriptomes.
Nature. 456:470–476. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ashley CT, Sutcliffe JS, Kunst CB, et al:
Human and murine FMR-1: Alternative splicing and translational
initiation downstream of the CGG-repeat. Nat Genet. 4:244–251.
1993. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Eichler EE, Richards S, Gibbs RA and
Nelson DL: Fine structure of the human FMR1 gene. Hum Mol Genet.
2:1147–1153. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Verheij C, Bakker CE, de Graaff E, et al:
Characterization and localization of the FMR-1 gene product
associated with fragile X syndrome. Nature. 363:722–724. 1993.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Verkerk AJ, de Graaff E, De Boulle K, et
al: Alternative splicing in the fragile X gene FMR1. Hum Mol Genet.
2:399–404. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sittler A, Devys D, Weber C and Mandel JL:
Alternative splicing of exon 14 determines nuclear or cytoplasmic
localisation of fmr1 protein isoforms. Human Mol Genet. 5:95–102.
1996. View Article : Google Scholar
|
|
8
|
Blencowe BJ: Alternative splicing: new
insights from global analyses. Cell. 126:37–47. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Huang T, Li LY, Shen Y, Qin XB, Pang ZL
and Wu GY: Alternative splicing of the FMR1 gene in human fetal
brain neurons. Am J Med Genet. 64:252–255. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ding J, Huang T, Li L, Fan Y and Shen Y:
Alternative splicing of the FMR1 gene in human fetal tissues.
Zhongguo Yi Xue Ke Xue Yuan Xue Bao. 19:241–246. 1997.In Chinese.
PubMed/NCBI
|
|
11
|
Thackeray JR and Ganetzky B:
Developmentally regulated alternative splicing generates a complex
array of Drosophila para sodium channel isoforms. J Neurosci.
14:2569–2578. 1994.PubMed/NCBI
|
|
12
|
Leparc GG and Mitra RD: A sensitive
procedure to detect alternatively spliced mRNA in pooled-tissue
samples. Nucleic Acids Res. 35:e1462007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kuhn RM, Haussler D and Kent WJ: The UCSC
genome browser and associated tools. Brief Bioinform. 14:144–161.
2013. View Article : Google Scholar :
|
|
14
|
Tamura K, Peterson D, Peterson N, Stecher
G, Nei M and Kumar S: MEGA5: Molecular evolutionary genetics
analysis using maximum likelihood, evolutionary distance, and
maximum parsimony methods. Mol Biol Evol. 28:2731–2739. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Poon AF, Frost SD and Pond SL: Detecting
signatures of selection from DNA sequences using Datamonkey.
Methods Mol Biol. 537:163–183. 2009.PubMed/NCBI
|
|
16
|
Pond SL and Frost SD: Datamonkey: Rapid
detection of selective pressure on individual sites of codon
alignments. Bioinformatics. 21:2531–2533. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Koscielny G, Le Texier V, Gopalakrishnan
C, et al: ASTD: The alternative splicing and transcript diversity
database. Genomics. 93:213–220. 2009. View Article : Google Scholar
|
|
18
|
Takeda J, Suzuki Y, Sakate R, et al:
H-DBAS: Human-transcriptome database for alternative splicing:
Update 2010. Nucleic Acids Res. 38:D86–D90. 2010. View Article : Google Scholar :
|
|
19
|
Ashley CT Jr, Wilkinson KD, Reines D and
Warren ST: FMR1 protein: Conserved RNP family domains and selective
RNA binding. Science. 262:563–566. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Santoro MR, Bray SM and Warren ST:
Molecular mechanisms of fragile X syndrome: A twenty-year
perspective. Annu Rev Pathol. 7:219–245. 2012. View Article : Google Scholar
|
|
21
|
Ascano M Jr, Mukherjee N, Bandaru P, et
al: FMRP targets distinct mRNA sequence elements to regulate
protein expression. Nature. 492:382–386. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Denman RB and Sung YJ: Species-specific
and isoform-specific RNA binding of human and mouse fragile X
mental retardation proteins. Biochem Biophys Res Commun.
292:1063–1069. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Xie W, Dolzhanskaya N, LaFauci G, Dobkin C
and Denman RB: Tissue and developmental regulation of fragile X
mental retardation 1 exon 12 and 15 isoforms. Neurobiol Dis.
35:52–62. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
De Boulle K, Verkerk AJ, Reyniers E, et
al: A point mutation in the FMR-1 gene associated with fragile X
mental retardation. Nat Genet. 3:31–35. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yang Z and Nielsen R: Estimating
synonymous and nonsynonymous substitution rates under realistic
evolutionary models. Mol Biol Evol. 17:32–43. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Xing Y and Lee C: Alternative splicing and
RNA selection pressure – evolutionary consequences for eukaryotic
genomes. Nat Rev Genet. 7:499–509. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Xing Y and Lee C: Evidence of functional
selection pressure for alternative splicing events that accelerate
evolution of protein subsequences. Proc Natl Acad Sci USA.
102:13526–13531. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Filip LC and Mundy NI: Rapid evolution by
positive Darwinian selection in the extracellular domain of the
abundant lymphocyte protein CD45 in primates. Mol Biol Evol.
21:1504–1511. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Xing Y and Lee CJ: Protein modularity of
alternatively spliced exons is associated with tissue-specific
regulation of alternative splicing. PLoS Genet. 1:e342005.
View Article : Google Scholar : PubMed/NCBI
|