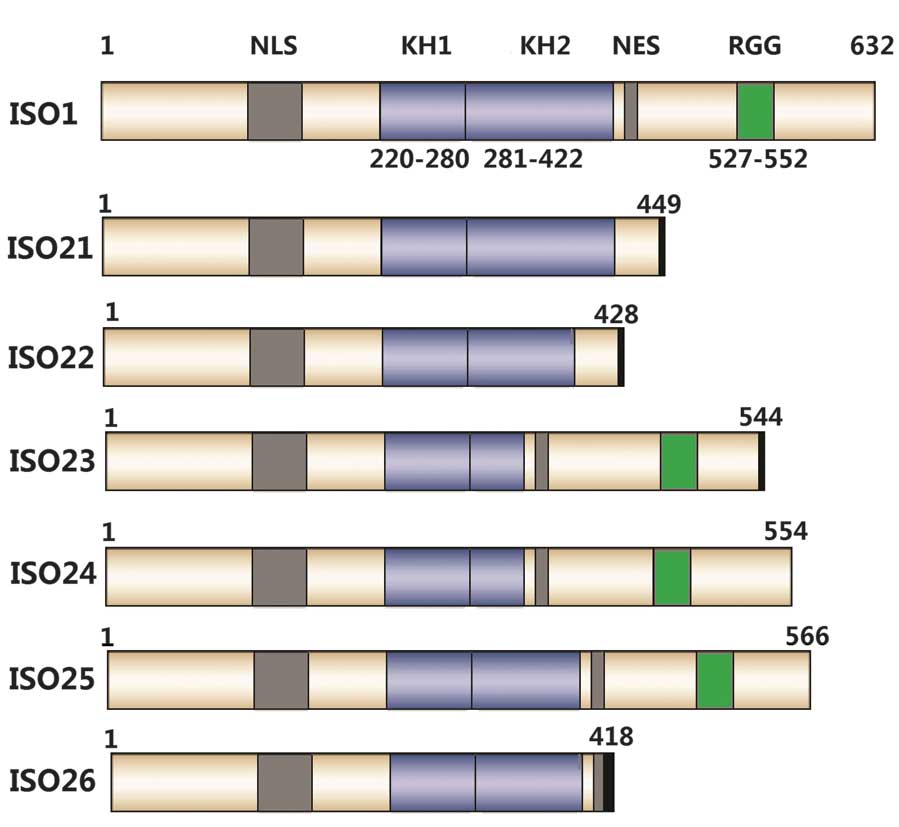
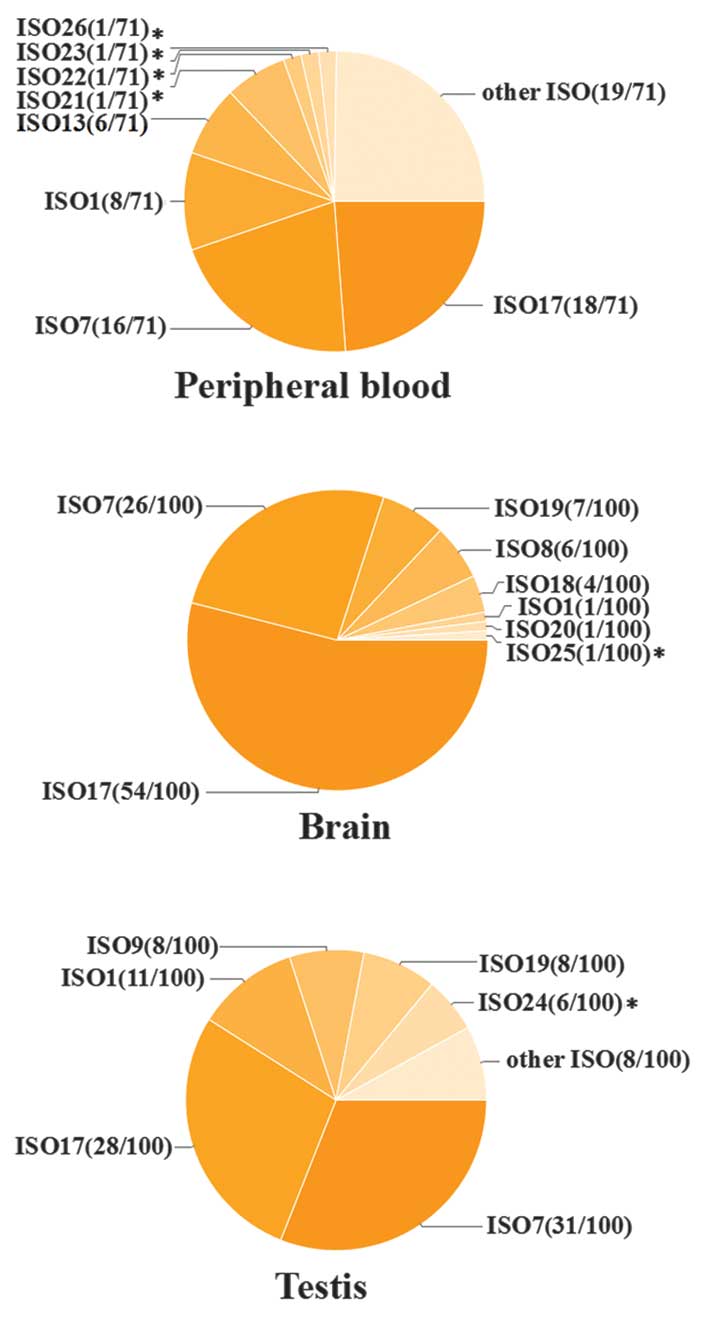
|
1
|
Johnson JM, Castle J, Garrett-Engele P, et
al: Genome-wide survey of human alternative pre-mRNA splicing with
exon junction microarrays. Science. 302:2141–2144. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wang ET, Sandberg R, Luo S, et al:
Alternative isoform regulation in human tissue transcriptomes.
Nature. 456:470–476. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ashley CT, Sutcliffe JS, Kunst CB, et al:
Human and murine FMR-1: Alternative splicing and translational
initiation downstream of the CGG-repeat. Nat Genet. 4:244–251.
1993. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Eichler EE, Richards S, Gibbs RA and
Nelson DL: Fine structure of the human FMR1 gene. Hum Mol Genet.
2:1147–1153. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Verheij C, Bakker CE, de Graaff E, et al:
Characterization and localization of the FMR-1 gene product
associated with fragile X syndrome. Nature. 363:722–724. 1993.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Verkerk AJ, de Graaff E, De Boulle K, et
al: Alternative splicing in the fragile X gene FMR1. Hum Mol Genet.
2:399–404. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sittler A, Devys D, Weber C and Mandel JL:
Alternative splicing of exon 14 determines nuclear or cytoplasmic
localisation of fmr1 protein isoforms. Human Mol Genet. 5:95–102.
1996. View Article : Google Scholar
|
|
8
|
Blencowe BJ: Alternative splicing: new
insights from global analyses. Cell. 126:37–47. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Huang T, Li LY, Shen Y, Qin XB, Pang ZL
and Wu GY: Alternative splicing of the FMR1 gene in human fetal
brain neurons. Am J Med Genet. 64:252–255. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ding J, Huang T, Li L, Fan Y and Shen Y:
Alternative splicing of the FMR1 gene in human fetal tissues.
Zhongguo Yi Xue Ke Xue Yuan Xue Bao. 19:241–246. 1997.In Chinese.
PubMed/NCBI
|
|
11
|
Thackeray JR and Ganetzky B:
Developmentally regulated alternative splicing generates a complex
array of Drosophila para sodium channel isoforms. J Neurosci.
14:2569–2578. 1994.PubMed/NCBI
|
|
12
|
Leparc GG and Mitra RD: A sensitive
procedure to detect alternatively spliced mRNA in pooled-tissue
samples. Nucleic Acids Res. 35:e1462007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kuhn RM, Haussler D and Kent WJ: The UCSC
genome browser and associated tools. Brief Bioinform. 14:144–161.
2013. View Article : Google Scholar :
|
|
14
|
Tamura K, Peterson D, Peterson N, Stecher
G, Nei M and Kumar S: MEGA5: Molecular evolutionary genetics
analysis using maximum likelihood, evolutionary distance, and
maximum parsimony methods. Mol Biol Evol. 28:2731–2739. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Poon AF, Frost SD and Pond SL: Detecting
signatures of selection from DNA sequences using Datamonkey.
Methods Mol Biol. 537:163–183. 2009.PubMed/NCBI
|
|
16
|
Pond SL and Frost SD: Datamonkey: Rapid
detection of selective pressure on individual sites of codon
alignments. Bioinformatics. 21:2531–2533. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Koscielny G, Le Texier V, Gopalakrishnan
C, et al: ASTD: The alternative splicing and transcript diversity
database. Genomics. 93:213–220. 2009. View Article : Google Scholar
|
|
18
|
Takeda J, Suzuki Y, Sakate R, et al:
H-DBAS: Human-transcriptome database for alternative splicing:
Update 2010. Nucleic Acids Res. 38:D86–D90. 2010. View Article : Google Scholar :
|
|
19
|
Ashley CT Jr, Wilkinson KD, Reines D and
Warren ST: FMR1 protein: Conserved RNP family domains and selective
RNA binding. Science. 262:563–566. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Santoro MR, Bray SM and Warren ST:
Molecular mechanisms of fragile X syndrome: A twenty-year
perspective. Annu Rev Pathol. 7:219–245. 2012. View Article : Google Scholar
|
|
21
|
Ascano M Jr, Mukherjee N, Bandaru P, et
al: FMRP targets distinct mRNA sequence elements to regulate
protein expression. Nature. 492:382–386. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Denman RB and Sung YJ: Species-specific
and isoform-specific RNA binding of human and mouse fragile X
mental retardation proteins. Biochem Biophys Res Commun.
292:1063–1069. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Xie W, Dolzhanskaya N, LaFauci G, Dobkin C
and Denman RB: Tissue and developmental regulation of fragile X
mental retardation 1 exon 12 and 15 isoforms. Neurobiol Dis.
35:52–62. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
De Boulle K, Verkerk AJ, Reyniers E, et
al: A point mutation in the FMR-1 gene associated with fragile X
mental retardation. Nat Genet. 3:31–35. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yang Z and Nielsen R: Estimating
synonymous and nonsynonymous substitution rates under realistic
evolutionary models. Mol Biol Evol. 17:32–43. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Xing Y and Lee C: Alternative splicing and
RNA selection pressure – evolutionary consequences for eukaryotic
genomes. Nat Rev Genet. 7:499–509. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Xing Y and Lee C: Evidence of functional
selection pressure for alternative splicing events that accelerate
evolution of protein subsequences. Proc Natl Acad Sci USA.
102:13526–13531. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Filip LC and Mundy NI: Rapid evolution by
positive Darwinian selection in the extracellular domain of the
abundant lymphocyte protein CD45 in primates. Mol Biol Evol.
21:1504–1511. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Xing Y and Lee CJ: Protein modularity of
alternatively spliced exons is associated with tissue-specific
regulation of alternative splicing. PLoS Genet. 1:e342005.
View Article : Google Scholar : PubMed/NCBI
|