Introduction
Hypertension is a major risk factor in
cardiovascular disease and end-stage renal damage, and increases
mortality rates worldwide (1).
Essential hypertension (EH) is affected by genetic and
environmental factors (2). Genetic
factors are estimated to be responsible for 30~50% of variations in
blood pressure (BP) levels (3).
Previous studies have identified multiple genetic loci associated
with BP or hypertension in various ethnic populations (4,5).
Epidemiological studies have reported that multiple environmental
factors are associated with risk of hypertension (6,7).
Aberrant epigenetic modifications, including DNA
methylation, may bridge the environmental and genetic contributing
factors. Gene-body methylation was found to be positively
correlated with gene expression (8). Although the functions of the
gene-body CpG island (CGI) remain to be elucidated, methylation of
the gene-body is frequently associated with active transcription in
humans and other animals (9).
Aberrant DNA methylation has been extensively investigated in the
context of the pathogenesis of multiple types of cancer, including
colorectal cancer (10), lung
cancer (11) and leukemia
(12). However, little evidence
has demonstrated an association between DNA methylation and the
risk of EH. A significant decline in global DNA methylation levels
are observed in patients with EH and the trend continues alongside
the progression of hypertension (13). In addition, altered global DNA
methylation in the placentas of patients with pre-eclampsia was
demonstrated to be associated with maternal hypertension (14). Aberrant DNA methylation of the
11β-HSD2, Adrb1 and ADD1 genes was
demonstrated to be associated with EH (15,16).
Hypertension and diabetes are two closely
associated, common diseases and their coexistence may increase the
risk of cardiovascular disease (17). GCK, a candidate gene for
type 2 diabetes (18), encodes
glucokinase, which is a key enzyme involved in glucose metabolism
(19,20). The association between EH and
GCK polymorphisms was previously disclosed in several
studies (21,22). GCK gene-body hypomethylation
was associated with the risk of coronary heart disease (23), which is closely correlated with EH
(24,25). The present study performed an
association study of GCK gene-body methylation with the risk
of EH, in order to assess whether GCK methylation is
associated with EH, and to examine the interactions between
GCK methylation and age, as well as clinical indicators.
Materials and methods
Sample collection
Samples from a total of 47 patients with EH and 47
age-matched control individuals were collected from the community
residents in Ningbo Baizhang Street Community Health Service Center
(Zhejiang, China) and the Seventh Hospital of Ningbo (Zhejiang,
China). The samples were collected from Han Chinese individuals,
who had been living in Ningbo for a minimum of three generations.
Hypertensive patients were defined according to the ‘gold standard’
(26). All the hypertensive
patients had received antihypertensive medication for >3 months,
or had at least three consecutive records of systolic blood
pressure (SBP) >140 mmHg and/or diastolic blood pressure (DBP)
>90 mmHg (26). Those patients
exhibiting SBP <120 mm Hg and DBP <80 mmHg, with no family
history of hypertension in their first degree relatives were
recruited as control individuals. The control individuals had not
received antihypertensive therapy. The ages of the control
individuals were matched with those of the patients with EH. None
of the control or hypertensive individuals had a history of
diabetes mellitus, secondary hypertension, myocardial infarction,
stroke, renal failure, drug abuse or other serious diseases. A
calibrated mercury sphygmomanometer with an adult-sized cuff was
used to measure blood pressure according to the standard
instructions recommended by the American Heart Association
(27). Blood pressure was measured
in supine position by two trained observers, with an interval of
≥10 min. Following a 12 h overnight fast, blood samples were
obtained from the antecubital vein using vacutainer tubes
containing EDTA (Hebei Chaoran Medical Instrument Company, Baoding,
China) and were stored at −80°C for DNA extraction. All experiments
were approved by the Ethics Committee of Ningbo University (Ningbo,
China) and written informed consent was obtained from all
subjects.
Biochemical analyses
A nucleic acid extraction analyzer (Lab-Aid 820;
Zeesan Biotech, Xiamen, China) was used to extract genomic DNA from
the peripheral blood samples. The concentration of extracted DNA
was measured using an ultramicro nucleic acid ultraviolet tester
(NanoDrop 1000; Thermo Fisher Scientific, Wilmington, DE, USA).
Plasma levels of cholesterol, triglyceride (TG), alanine
transaminase (ALT), aspartate transaminase, uric acid and glucose
concentrations were enzymatically measured using a CX7 biochemical
analyzer (Beckman Coulter, Brea, CA, USA). DNA meth-ylation was
measured using sodium bisulphite DNA conversion coupled with
pyrosequencing (28). Briefly,
genomic DNA was chemically modified by sodium bisulphite (EpiTech
Bisulphite kit; Qiagen, Hilden, Germany) to convert all the
unmethylated cytosines into uracils, while the methylated cytosines
remained unchanged. The converted DNA were selected and the
polymerase chain reaction (PCR) primers were designed with PyroMark
Assay Design software, version 2.0.1.15 (Qiagen). The PCR products
were subsequently degenerated using dena-turation solution (Qiagen)
and released to single strand products for pyrosequencing (29). PCR amplification was performed in
reaction mixtures containing 10 µl ZymoTaq™ Premix (Zymo
Research Corporation, Irvine, California, USA), 1.5 µl
forward primer, 1.5 µl reverse primer, 2 µl converted
DNA and 5 µl DNAase/RNAase free water. PCR amplification was
conducted using an Eppendorf Mastercycler Nexus Gradient
(Eppendorf, Hamburg, Germany) under the following conditions: 95°C
for 10 min, then 40 cycles of 95°C for 30 sec, 54.5°C for 40 sec
and 72°C for 50 sec, followed by one cycle of 72°C for 7 min. The
primer sequences were as follows: Forward,
5′-TGGATGGTTTAGTGTATAAGTTGTATT-3′; reverse, 5′-Biotin-CACCTCATCCTCC
ACATTCAT-3′ and sequencing primer, 5′-AAGTGGGGTTTAAAAAG-3′.
Statistical analyses
Statistical analyses were performed to investigate
the association of GCK methylation with metabolic profile
and EH. A two sample t-test was performed to determine the
association of EH with continuous variables, including age, body
mass index, cholesterol, TG, glucose, ALT and uric acid. Pearson’s
correlation was used to determine the association between
GCK methylation and the biochemical indicators. A more
conservative non-parametric approach was used for data which were
unable to be normalized. Pearson’s χ2 or Fisher’s exact
test was used to evaluate the association between EH and
categorical variables, including gender, smoking and drinking. A
receiver operating characteristic curve was used to examine the
sensitivity of GCK methylation in EH diagnosis. In addition,
logistic regression was implemented to investigate the interactions
between GCK methylation and age. P<0.05 was considered to
indicate a statistically significant difference. The aforementioned
statistical analyses were performed using PASW Statistics 19.0
software [SPSS (Honk Kong) Ltd., Hong Kong, China].
Results
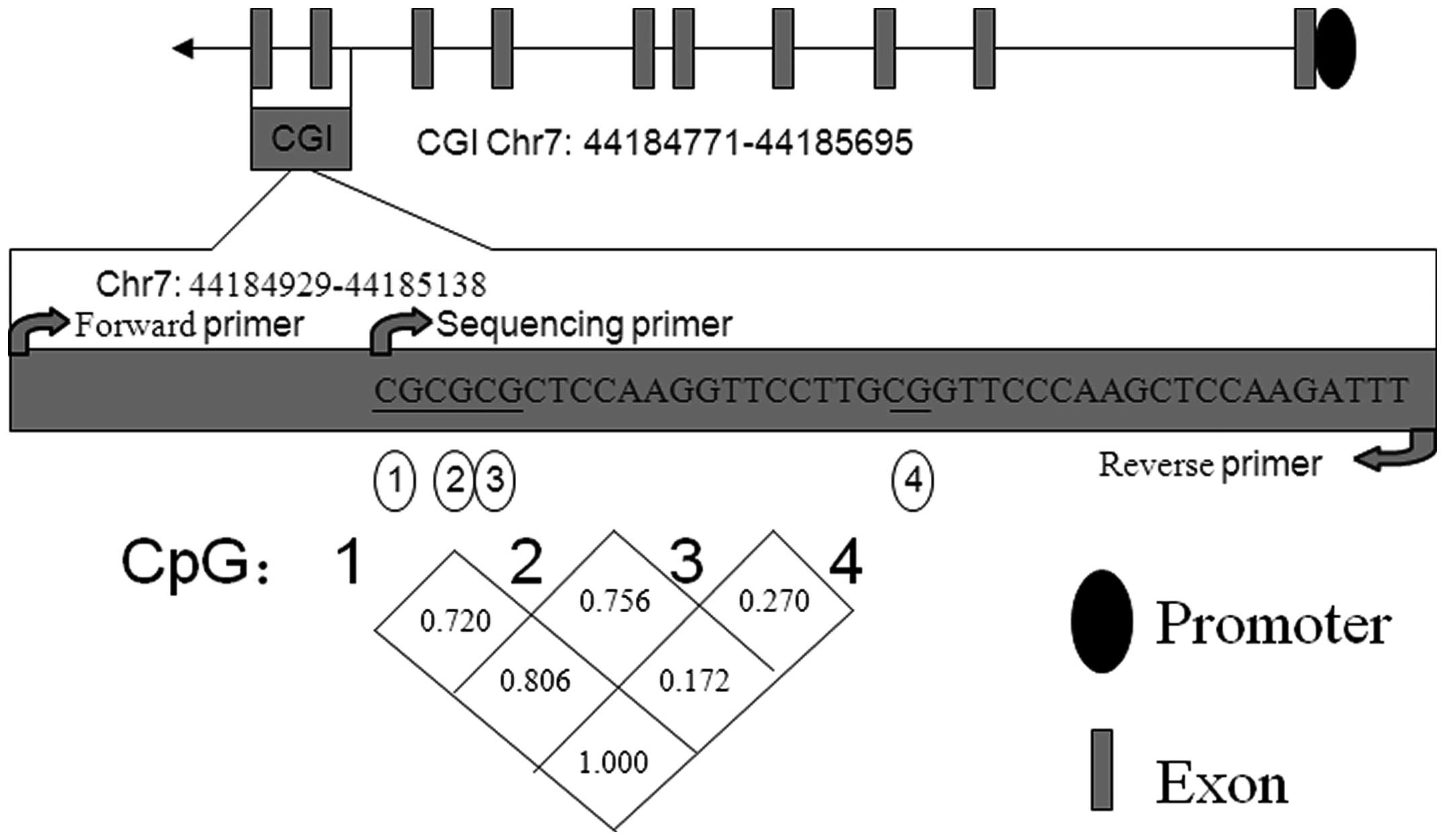
In the present study, a fragment in the GCK
gene-body CGI, spanning exon 9 and a section of exon 10 (hg19,
chr7: 44184771-44185695) was selected automatically by PyroMark
Assay Design software for analysis. The results demonstrated that
DNA methylation levels were closely correlated between CpG1, CpG2
and CpG3 (Fig. 1; r>0.70;
P<0.001). By contrast a markedly weaker correlation was observed
between CpG4 and the preceding three CpGs (r<0.3 or r=1;
P>0.05).
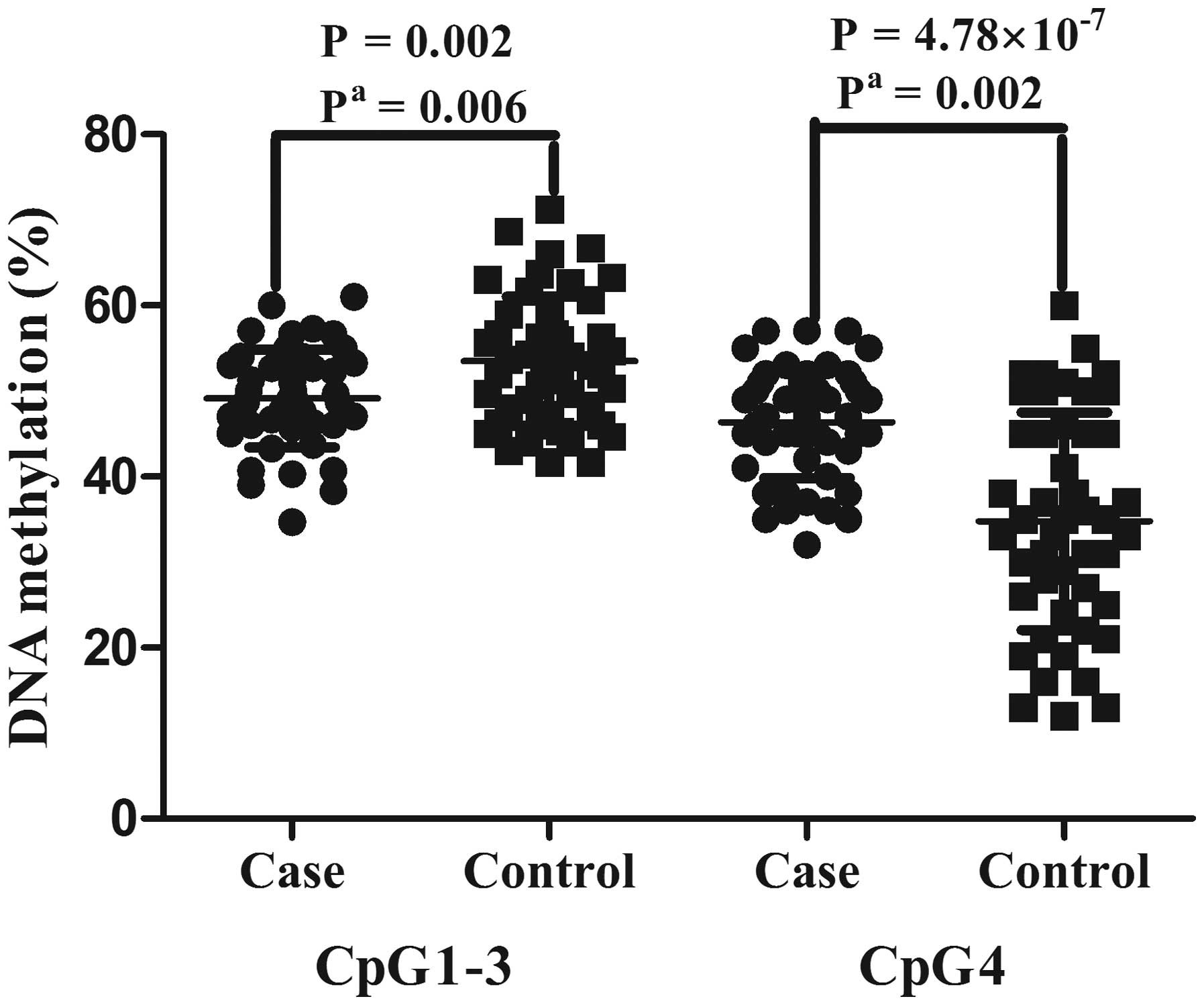
As shown in Table I
and Fig. 2, the results
demonstrated that CpG1-3 methylation levels were significantly
lower in samples from patients with EH (cases vs. controls,
49.13±5.72 vs. 53.49±7.53%; adjusted P=0.006). By contrast, the
CpG4 methylation levels were significantly higher in the samples
from patients with EH (cases vs. controls,46.34±6.48 vs.
34.74±12.73%; adjusted P=0.002). No correlation was detected
between GCK DNA methylation and age amongst the patients or
controls (data not shown), and no significant interactions of
GCK methylation and age were revealed to affect EH
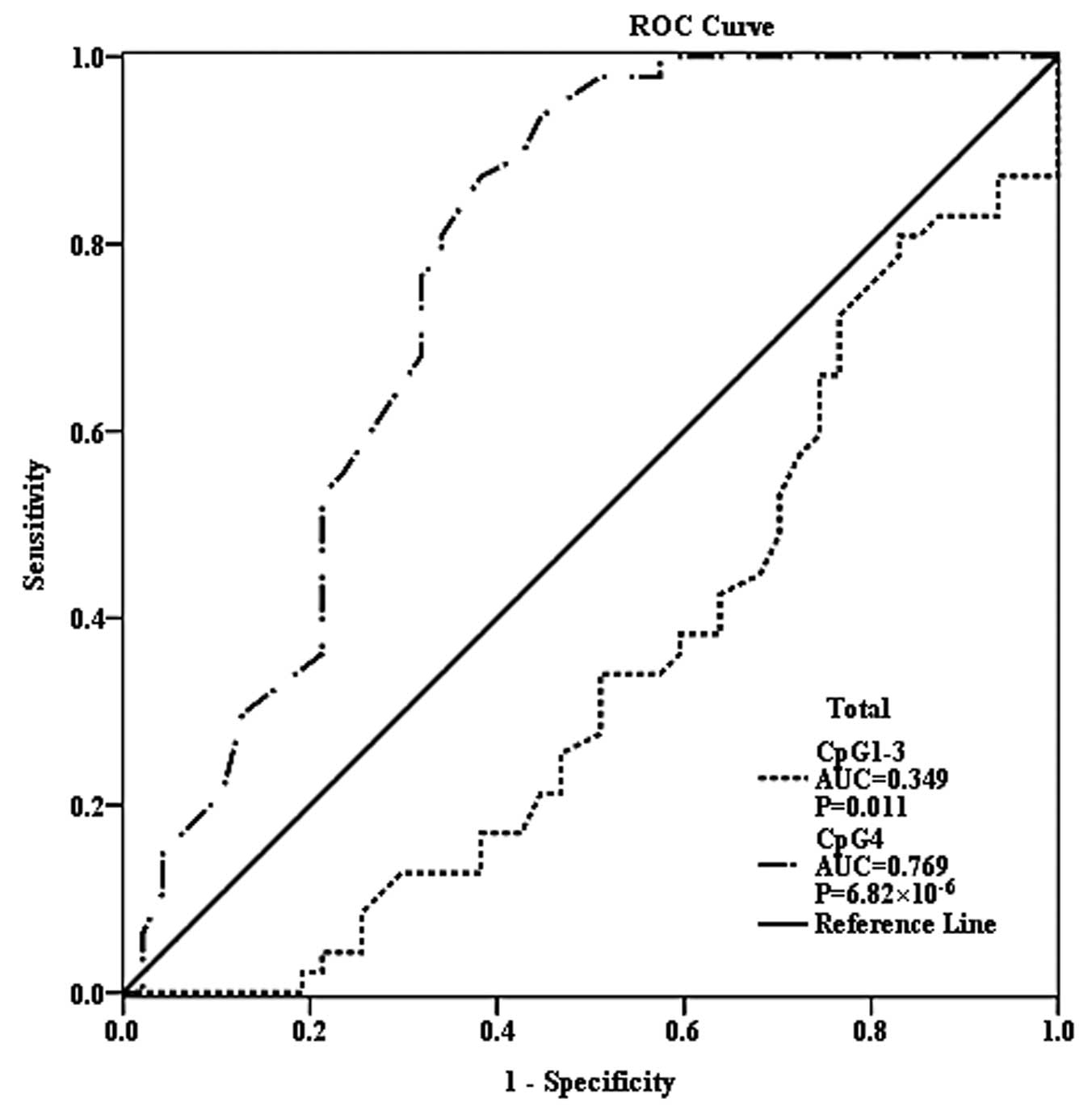
susceptibility (P>0.05). Additionally, CpG4 methylation level
was demonstrated to be an effective predictor of EH (Fig. 3; area under curve=0.769;
P=6.82×10−6), however, CpG1-3 methylation was not a
successful diagnostic marker of EH risk.
 | Table ICharacteristics of subjects. |
Table I
Characteristics of subjects.
| Characteristic | EH (Mean ± SD) | Non-EH (Mean ±
SD) | P-value |
|---|
| Age (years) | 59.34±7.20 | 59.21±7.62 | 0.934 |
| BMI
(kg/m2)a | 24.02±2.62 | 22.52±4.08 | 0.103 |
| Smoking (Y/N) | 7/40 | 8/27 | 0.356 |
| Alcoholic intake
(Y/N) | 16/31 | 5/30 | 0.043 |
| Cholesterol
(mmol/l) | 5.28±0.92 | 5.04±1.07 | 0.238 |
| TG (mmol/l) | 1.87±1.40 | 1.31±0.83 | 0.006 |
| Glucose
(mmol/l) | 5.43±0.52 | 5.23±0.92 | 0.001 |
| ALT (IU/l) | 26.26±18.58 | 18.17±12.14 | 0.007 |
| Uric acid
(mmol/l) | 363.71±84.82 | 302.00±91.30 | 0.001 |
| CpG1-3 methylation
(%) | 49.13±5.72 | 53.49±7.53 | 0.002 |
| CpG4 methylation
(%) | 46.34±6.48 | 34.74±12.73 |
4.78×10−7 |
As shown in Table
I, significant differences were observed in glucose (P=0.001),
TG (P=0.006), ALT (P=0.007) and uric acid (P=0.001) between EH
cases and controls. In addition, gender-stratified correlation
assessments were performed to assess the association between
GCK DNA methylation levels and metabolic phenotypes,
including glucose, triglycerides, cholesterol, uric acid and ALT,
in the control samples. A significant correlation between the
levels of CpG1-3 methylation and ALT (Fig. 4; r=−0.424; P=0.044) was observed in
females, while no significant results were obtained for the
remaining tests.
Discussion
Previous studies have reported that GCK
polymorphisms were associated with hypertension (21,22).
The present study hypothesized that the aberrant GCK
gene-body methylation may also induce EH. The results revealed that
patients with EH exhibited hypomethylation of CpG1-3 and
hypermethylation of CpG4 in the GCK gene-body, compared with
those of the control individuals. The results of the present study
regarding the role of GCK methylation in the risk of EH may
provide novel hints to aid the clarification of the pathogenesis of
EH in future.
In the present study, no specific CGI was observed
in the human GCK promoter. Gene-body methylation identified
‘orphan promoters’, which may be used in the early stages of
development (30).
Hypermethylation of gene-bodies frequently indicates higher levels
of gene expression in human tissues and cell types (8,31).
Aberrant methylation of the gene body was observed to contribute to
the risk of heart failure (32)
and coronary heart disease (23).
DNA methylation of gene bodies has major significance in the
regulation of gene expression (33). In certain types of human tissue,
positive correlation between gene-body methylation and gene
expression has been identified (34,35).
Gene-body methylation was reported to be significant in regulating
cell context-specific alternative promoters in human and mouse
tissues (28).
Associations between DNA methylation and age were
demonstrated to affect the EH status (16). This observation indicated that EH
was affected by genetic and environmental factors, including DNA
methylation status and age. Age was previously revealed to be
positively correlated with GCK methylation levels (23), however the present study failed to
demonstrate this association. No association between age and CpG1-3
or CpG4 methylation was observed (data not shown).
A positive correlation between plasma glucose levels
and blood pressure has been previously reported (36). Increased levels of plasma ALT
(37) and uric acid (38) were revealed to be associated with
hypertension. In the present study, the association of uric acid
and plasma glucose with GCK DNA methylation levels was not
observed, whereas CpG1-3 methylation levels were positively
associated with ALT in the female control samples. These results
suggested that GCK CpG1-3 methylation may exert its effects
on the risk of EH via regulation of the levels of ALT in females.
Ongoing research is required to investigate the specific mechanisms
underlying this association.
In conclusion, the results of the present study
demonstrated that a hypomethylation level of CpG1-3 and a
hypermethylation level of CpG4 in the GCK gene-body
increased the risk of EH. GCK CpG4 methylation may have
potential in aiding the prediction of EH risk. The findings provide
novel clues to facilitate the elucidation of the pathogenesis of
EH.
Acknowledgments
This study was supported by grants from the National
Natural Science Foundation of China (no. 81373094), Zhejiang
Provincial Natural Science Foundation of China (no. Y2100857), the
K.C. Wong Magna Fund in Ningbo University, Natural Science
Foundation of Ningbo City (no. 2011A610037) and the Research
Project of Ningbo University (no. XKL11D2124).
References
|
1
|
Lawes CM, Vander Hoorn S and Rodgers A;
International Society of Hypertension: Global burden of
blood-pressure-related disease, 2001. Lancet. 371:1513–1518. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Xi B, Cheng H, Shen Y, et al: Physical
activity modifies the associations between genetic variants and
hypertension in the Chinese children. Atherosclerosis. 225:376–380.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Tanira MO and Al Balushi KA: Genetic
variations related to hypertension: A review. J Hum Hypertens.
19:7–19. 2005. View Article : Google Scholar
|
|
4
|
Xi B, Shen Y, Reilly KH, Wang X and Mi J:
Recapitulation of four hypertension susceptibility genes (CSK,
CYP17A1, MTHFR and FGF5) in East Asians. Metabolism. 62:196–203.
2013. View Article : Google Scholar
|
|
5
|
Xi B, Tang W and Wang Q: Polymorphism near
the ATP2B1 gene is associated with hypertension risk in East
Asians: A meta-analysis involving 15909 cases and 18529 controls.
Blood Press. 21:134–138. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Binder A: A review of the genetics of
essential hypertension. Curr Opin Cardiol. 22:176–184. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Whelton PK, He J, Appel LJ, et al: Primary
prevention of hypertension: Clinical and public health advisory
from The National High Blood Pressure Education Program. JAMA.
288:1882–1888. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Jjingo D, Conley AB, Yi SV, Lunyak VV and
Jordan IK: On the presence and role of human gene-body DNA
methylation. Oncotarget. 3:462–474. 2012.PubMed/NCBI
|
|
9
|
Zemach A, McDaniel IE, Silva P and
Zilberman D: Genome-wide evolutionary analysis of eukaryotic DNA
methylation. Science. 328:916–919. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kim MS, Lee J and Sidransky D: DNA
methylation markers in colorectal cancer. Cancer Metastasis Rev.
29:181–206. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kerr KM, Galler JS, Hagen JA, Laird PW and
Laird-Offringa IA: The role of DNA methylation in the development
and progression of lung adenocarcinoma. Dis Markers. 23:5–30. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Akalin A, Garrett-Bakelman FE, Kormaksson
M, et al: Base-pair resolution DNA methylation sequencing reveals
profoundly divergent epigenetic landscapes in acute myeloid
leukemia. PLoS Genet. 8:e10027812012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Smolarek I, Wyszko E, Barciszewska AM, et
al: Global DNA methylation changes in blood of patients with
essential hypertension. Med Sci Monit. 16:CR149–CR155.
2010.PubMed/NCBI
|
|
14
|
Kulkarni A, Chavan-Gautam P, Mehendale S,
Yadav H and Joshi S: Global DNA methylation patterns in placenta
and its association with maternal hypertension in pre-eclampsia.
DNA Cell Biol. 30:79–84. 2011. View Article : Google Scholar
|
|
15
|
Friso S, Pizzolo F, Choi SW, et al:
Epigenetic control of 11 beta-hydroxysteroid dehydrogenase 2 gene
promoter is related to human hypertension. Atherosclerosis.
199:323–327. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhang LN, Liu PP, Wang L, et al: Lower
ADD1 gene promoter DNA methylation increases the risk of essential
hypertension. PLoS One. 8:e634552013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liu J, Zhao D, Liu J, Qi Y, Sun J and Wang
W: Prevalence of diabetes mellitus in outpatients with essential
hypertension in China: A cross-sectional study. BMJ Open.
3:e0037982013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Iynedjian PB: Molecular physiology of
mammalian glucokinase. Cell Mol Life Sci. 66:27–42. 2009.
View Article : Google Scholar :
|
|
19
|
Heredia VV, Thomson J, Nettleton D and Sun
S: Glucose-induced conformational changes in glucokinase mediate
allosteric regulation: Transient kinetic analysis. Biochemistry.
45:7553–7562. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kamata K, Mitsuya M, Nishimura T, Eiki Ji
and Nagata Y: Structural basis for allosteric regulation of the
monomeric allo-steric enzyme human glucokinase. Structure.
12:429–438. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chiang FT, Chiu KC, Tseng YZ, Lee KC and
Chuang LM: Nucleotide(-258) G-to-A transition variant of the liver
glucokinase gene is associated with essential hypertension. Am J
Hypertens. 10:1049–1052. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yamada Y, Ando F and Shimokata H:
Association of polymorphisms of SORBS1, GCK and WISP1 with
hypertension in community-dwelling Japanese individuals. Hypertens
Res. 32:325–331. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Xu L, Zheng D, Wang L, et al: GCK
gene-body hypomethylation is associated with the risk of coronary
heart disease. Biomed Res Int. 2014:72014. View Article : Google Scholar
|
|
24
|
MacMahon S, Peto R, Collins R, et al:
Blood pressure, stroke and coronary heart disease: Part 1,
prolonged differences in blood pressure: prospective observational
studies corrected for the regression dilution bias. Lancet.
335:765–774. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Nadeem M, Ahmed SS, Mansoor S and Farooq
S: Risk factors for coronary heart disease in patients below 45
years of age. Pak J Med Sci. 29:91–96. 2013.PubMed/NCBI
|
|
26
|
European Society of Hypertension-European
Society of Cardiology Guidelines Committee: 2003 European Society
of Hypertension-European Society of Cardiology guidelines for the
management of arterial hypertension. J Hypertens. 21:1011–1053.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Perloff D, Grim C, Flack J, et al: Human
blood pressure determination by sphygmomanometry. Circulation.
88:2460–2470. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Jiang D, Zheng D, Wang L, et al: Elevated
PLA2G7 gene promoter methylation as a gender-specific marker of
aging increases the risk of coronary heart disease in females. PLoS
One. 8:e597522013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Bassil CF, Huang Z and Murphy SK:
Bisulfite pyrosequencing. Methods Mol Biol. 1049:95–107.
2013.PubMed/NCBI
|
|
30
|
Illingworth RS, Gruenewald-Schneider U,
Webb S, et al: Orphan CpG islands identify numerous conserved
promoters in the mammalian genome. PLoS Genet. 6:e10011342010.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Maunakea AK, Nagarajan RP, Bilenky M, et
al: Conserved role of intragenic DNA methylation in regulating
alternative promoters. Nature. 466:253–257. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Movassagh M, Choy MK, Knowles DA, et al:
Distinct epigenomic features in end-stage failing human hearts.
Circulation. 124:2411–2422. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Jones PA: Functions of DNA methylation:
Islands, start sites, gene bodies and beyond. Nat Rev Genet.
13:484–492. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
34
|
Aran D, Toperoff G, Rosenberg M and
Hellman A: Replication timing-related and gene body-specific
methylation of active human genes. Hum Mol Genet. 20:670–680. 2011.
View Article : Google Scholar
|
|
35
|
Ball MP, Li JB, Gao Y, et al: Targeted and
genome-scale strategies reveal gene-body methylation signatures in
human cells. Nat Biotechnol. 27:361–368. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Cambien F, Warnet JM, Eschwege E,
Jacqueson A, Richard JL and Rosselin G: Body mass, blood pressure,
glucose, and lipids. Does plasma insulin explain their
relationships? Arteriosclerosis. 7:197–202. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Cheung BM, Ong KL, Tso AW, et al:
Gamma-glutamyl trans-ferase level predicts the development of
hypertension in Hong Kong Chinese. Clin Chim Acta. 412:1326–1331.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Chen DL, Zhang CJ, Fu YH, Mo YJ and Chen
FR: Correlation of angiotensin-converting enzyme 2 gene
polymorphisms to essential hypertension and ischemic stroke. Nan
Fang Yi Ke Da Xue Xue Bao. 30:1890–1892. 2010.In Chinese.
|