Introduction
Retinal injury is one of most common eye diseases
and is associated with old age, light damage and trauma (1–3).
When eyes are directly and chronically exposed to sources of light
(4–6), such as the Sun and lasers, the retina
may be irreversibly damaged. Visible light (400–760 nm) may damage
the retina through photochemical injury (7–9),
which is considered to be the most common mechanism of
light-induced retinal injury. Photochemical injury is independent
of either mechanical or thermal retinal damage, and was first
documented by Noell et al in 1966 (10). The authors showed that the retinas
of albino rats were irreversibly damaged by continuous exposure to
ambient light within the range of the visible light spectrum.
Photochemical retinal-injury may be categorized into two distinct
classes (11). Class I injury is
characterized by exposure to low levels of white light irradiance
(<1 mW/cm2) over hours to weeks. Class II injury is
characterized by exposure to high levels of white light irradiance
(>10 mW/cm2), with an action spectrum at shorter
wavelengths. Rattner et al (12) found an increase in RNA transcripts
coding for protective proteins, such as matrix metalloproteinase-3,
serine peptidase inhibitor, clade A, member 3N; Serpin b1a; and the
oncostatin M receptor. Rattner et al (12) also demonstrated a decrease in the
transcription of genes coding for visual cycle components in
neurosensory and retinal pigment epithelial (RPE) cells in response
to light, using microarray RNA blotting and in situ
hybridization (12).
MicroRNA (miRNA; ~22 nucleotides) is a type of
small, endogenous and non-coding regulatory RNA, which is involved
in the downregulation of target genes in a number of organisms,
such as microorganisms and mammals (13–16).
A study by Lee et al (17),
demonstrated that lin-4 and let-7 in
Caenorhabditis elegans caused phenotypic functional loss,
and their mutation prevented them from developing into larval
stage. Since then, numerous studies have identified miRNA in
organisms, including Drosophila (18,19)
and mammals (20,21). miRNA binds to complementary
sequences in the 3′-untranslated region (UTR) of multiple target
mRNAs, usually inhibiting translation or causing destabilization
(22,23). Certain miRNAs, such as the
hsa-miR-183/96/182 cluster and hsa-miR-220, are able to silence
retinal-specific genes, which are predominantly expressed in the
retina (24–26). Xu et al (27) reported that in mouse embryonic
retinas, hsa-miR-183/96/182 expression is minimal, whereas its
expression increases markedly following birth and peaks in adult
retinas. hsa-miR-183/96/182 expression has a function in the
maturation and healthy functioning of the adult retina (27). Krol et al (28) demonstrated that hsa-miR-183/96/182
exhibits a response to light and rapid turnover as a common
property of neuronal microRNAs, which is independent of the
circadian cycle. Lumayag et al (29) showed that inactivation of the
hsa-miR-183/96/182 cluster results in changes in retinal gene
expression, postnatal functional differentiation and synaptic
connectivity of photoreceptors. A study by Liu et al’s
(25) demonstrated that synthetic
miRNA-mowers, targeting the has-miR-183/96/182 cluster, are
involved in bladder cancer development. However, to the best of our
knowledge, the influence of the hsa-miR-183/96/182 cluster on the
downregulation of brain-derived neurotrophic factor (BDNF) in the
retina has not been investigated. Therefore, the aim of the present
study was to investigate the expression of hsa-miR-183/96/182
cluster following light treatment and to further investigate its
regulation on the BDNF gene.
Materials and methods
Culturing human primary RPE cells and
experimental grouping
The human primary cultured RPE cells (Beijing
Research Biotechnology Co., Ltd. Beijing, China) were resurrected
and cultured in α modified Eagle’s medium (Thermo Fisher Scientific
Co., Ltd., Waltham, MA, USA) with 2 mM L-glutamine (Beijing
Beifangtongzheng Co., Ltd., Beijing, China), 100 U/ml penicillin
(Beijing Beifangtongzheng Co., Ltd.), 100 mg/ml streptomycin
(Beijing Beifangtongzheng Co., Ltd.) and 10% fetal bovine serum
(Thermo Fisher Scientific Co., Ltd.). Following this, the RPE cells
were incubated at 37°C in 5% CO2 for one week, and
randomly divided into two groups when cells were planked with 80%.
The RPE cells in the visible light exposure group were exposed to
visible light (400–760 nm) for 4 h, while the RPE cells in the dark
condition group were shaded with silver paper for 4 h.
Subsequently, cells were harvested in order to separate total RNA
for reverse transcription-quantitative polymerase chain reaction
(RT-qPCR). Experiments for each group were repeated at least three
times.
The potential target genes of the has-miR-183/96/182
cluster were identified using TargetScan software (http://www.targetscan.org/) according to the
manufacturer’s instructions.
Synthesis of hsa-miR-183-96-182 mimics
and transfection of the RPE cells
The hsa-miR-183, hsa-miR-96, hsa-miR-182 and
has-miR-183/96/182 mimics were designed according to the sequence
from the miRBase website (http://www.mirbase.org/). They were artificially
synthesized by Invitrogen Life Technologies (Carlsbad, CA, USA).
RPE cells were inoculated on a 12-well plate for 24 h prior to
transfection, when planked with 80% and then transfected into RPE
cells with 1 μg of each vector using lipofectamine
2000® transfection reagent (Invitrogen Life
Technologies), according to the manufacturer’s instructions.
Following 24 h transfection, cells were harvested to separate total
RNA and total protein for subsequent experiments. Experiments for
each group were repeated at least three times.
RT-qPCR
Total RNA from the RPE cells was extracted using a
miRcute miRNA isolation kit (Tiangen Biotech, Beijing, China)
according to the manufacturer’s instructions. The RPE cells
(1×108/cm3) were harvested and disrupted by
buffer MZ at room temperature (RT) for 5 min. Chloroform (200
μl) was added to the supernatant, followed by vigorous
shaking for 15 sec and incubation at RT for 5 min. Following this,
the cells were centrifuged at 12,000 × g at 4°C for 15 min. The
upper aqueous phase was transferred to a 2 ml collection tube, and
1.5X volumes of anhydrous ethanol (Beijing Beifangtongzheng Co.,
Ltd.) was added followed by thorough mixing. The sample (700
μl) was added into a miRelute column and centrifuged at
12,000 × g at RT for 30 sec in order to discard the flow-through.
MRD buffer (500 μl) was added to the miRelute column,
incubated at RT for 2 min and centrifuged at 12,000 × g at RT for
30 sec to discard the flow-through. RW buffer (700 μl) was
added to the miRelute column, incubated at RT for 2 min and then
centrifuged at 12,000 × g at RT for 30 sec in to wash the column
and repeated again. The miRelute column was placed into a new
collection tube and centrifuged at 12,000 × g at 4°C for 1 min in
order to dry the miRspin column. RNase-free ddH2O
(30–100 μl; Beijing Beifangtongzheng Co., Ltd.) was added
directly onto the miRspin column, incubated at RT for 2 min, and
centrifuged at 12,000 × g at RT for 2 min in order to elute total
RNA.
In order to conduct an RT-qPCR assay of the
hsa-miR-183/96/182 cluster in RPE cells, total RNA was
reverse-transcribed and analyzed using a SYBR®
PrimeScript™ miRNA qPCR kit (Takara Bio, Inc., Shiga, Japan)
according to the manufacturer’s instructions. The following primers
were used in order to amplify the hsa-miR-183, hsa-miR-96 and
hsa-miR-182 in the RPE cells after visible light exposure: Forward:
5′-ACACTCCAGCTGGGTATGGCACTGGTAGAA-3′ and reverse:
5′-CTCAACTGGTGTCGTGGAGTCG-GCAATTCAGTTGAG AGUGAAUU-3′ for
hsa-miR-183, forward: 5′-ACACTCCAGCTGGGTTTGGCACTAGCACATT-3′ and
reverse: 5′-CTCAACTGGTGTCGTGGAGTCGGCA ATTCAGTTGAGAGCAAAAA-3′ for
hsa-miR-9, forward: 5′-ACACTCCAGCTGGGTTTGGCAATGGTAGAACT-3′ and
reverse: 5′-CTCAACTGGTGTCGTGGAGTCGGCA ATTCAGTTGAGAGUGUGAG-3′ for
hsa-miR-182, and forward: 5′-CTCGCTTCGGCAGCACA-3′ and reverse:
5′-AACGCTTCACGAATTTGCGT-3′ for U6.
In order to conduct an RT-qPCR assay of BDNF in RPE
cells, total RNA was reverse-transcribed using a total RNA kit II
(Omega-Biotek, Norcross, GA, USA) according to the manufacturer’s
instructions. Amplification was conducted using SYBR®
Premix Ex Taq™ II system (Takara Bio, Inc.) using the following
primers: Forward: 5′-CTACGAGACCAAGTGCAATCC-3′ and reverse:
5′-AATCGCCAGCCAATTCTCTTT-3′ for BDNF.
All PCR reactions consisted of a 20 μl
solution of 10 μl SYBR Green PCR Master Mix (Takara Bio,
Inc., Shiga, Japan), 2 μl forward primer, 2 μl
reverse primer, 1 μl template and 5 μl RNase-free
water in a 0.2 ml EP tube, and used the following PCR protocol:
95°C for 15 min, followed by 45 cycles of 94°C for 15 sec, 55°C for
30 sec and 70°C for 30 sec. The relative quantification of miRNA
was calculated using the 2−ΔΔCT method.
Western blot analysis
Protein concentration was measured using the
bicinchoninic acid protein assay reagent (Thermo Fisher Scientific
Inc., Waltham, MA, USA) and. Samples (~35 μg) were separated
using electrophoresis through 15% polyacrylamide gels, and
transferred to polyvinylidene difluoride membranes (GE Healthcare,
Cleveland, OH, USA) according to the manufacturer’s instructions.
The membranes were probed with mouse-derived anti-BDNF monoclonal
antibody (Beijing Zhongshan Golden Bridge Biotechnology, Co., Ltd.,
Beijing, China) diluted to 1:1,000 in Tris-Buffered Saline with
Tween-20 (TBST; Sigma-Aldrich, St. Louis, MO, USA) and
mouse-derived anti-β-actin antibody diluted to 1:1,000 in TBST
(Beijing Zhongshan Golden Bridge, Biotechnology, Co., Beijing
China) for 1.5 h at RT, followed by a horseradish
peroxidase-conjugated goat anti-mouse secondary antibody (1:5,000
in TBST; Beijing Zhongshan Golden Bridge Biotechnology, Co., Ltd.)
and incubated at RT for 1 h. The immunolabeled bands were examined
by exposure to an X-ray film using the chemiluminescent substrate
luminal reagent (GE Healthcare). The optical density of the band
was scanned and quantified using the ImageJ software version 1.46
(http://www.originlab.com/).
Construction of psiCHECK™-2-BDNF (wild
type) and psiCHECK-2-BDNF (mutation)
The BDNF mutant was designed based on the putative
miR-182 recognition site (located at 252–258 bp) in the BDNF
3′-UTR, and mutated using QuikChange Lightning Site-Directed
Mutagenesis kit (Stratagene, La Jolla, CA, USA) according to the
manufacturer’s instructions. The BDNF cDNA encoding sequences,
including wild type and mutant, were amplified using PCR of human
genomic DNA, using the following primers: 5′-CTCGAGACATGTCCATGACCAGAAGGG-3′
(underline indicates the Xho I restriction endonuclease site
and 5′-GCGGCCGCTCACAATTAAAGCAGCATGCAA-3′
(underline indicates the Not I restriction endonuclease
site. Subsequently, the PCR products and psiCHECK-2 vector were
digested using restriction endonucleases Xho I (Takara Bio,
Inc.) and Not I (Takara Bio, Inc.), and conjugated using
T4 DNA ligase (Takara Bio, Inc.), followed by direct
sequencing by Invitrogen Life Technologies.
Luciferase activity assay
In order to conduct a dual luciferase
(Renilla luciferase/firefly luciferase) reporter gene assay,
the 293T cells (Beijing Research Biotechnology Co., Ltd.) were
cultured with Dulbecco’s modified Eagle’s medium, inoculated on a
12 well plate, and divided into four groups: psiCHECK-2, BDNF (wild
type); mimic + BDNF (wild type); and mimic + BDNF (mutant). When
the 293T cells were planked with 80%, 1 μg of each device
was then transfected into 293T cells using Lipofectamine 2000
transfection reagent according to the manufacturer’s instructions.
Following 48 h transfection, the 293T cells were harvested and
digested with 0.25% trypsin (Takara Bio, Inc.). The 293T cells (75
μl; 5×104/ml) were separated and equal volumes of
luciferase reagent was added to the cells for lysis at RT for 10
min, following which the firefly luciferase activity was measured
Subsequently, 75 μl dual luciferase was added in order to
fix the reagent to the plate and lysed at RT for 10 min, and then
the Renilla luciferase activity was measured. The ratio of
Renilla luciferase/firefly luciferase was then calculated
and a histogram was produced using Origin 9.0 software (http://www.originlab.com/).
Statistical analysis
The data were analyzed by SPSS 21.0 (IBM SPSS,
Armonk, NY, USA) using Student’s t-test. P<0.05 was considered
to indicate a statistically significant difference.
Results
Visible light exposure increased
hsa-miR-183/96/182 cluster expression in the RPE cells
hsa-miR-183, hsa-miR-96 and hsa-miR-182
transcription increased significantly in the RPE cells exposed to
visible light compared with that in the RPE cells exposed to dark
conditions (Fig. 1A, P<0.01).
The potential target genes of the hsa-miR-183/96/182 cluster in the
RPE cells were predicted using TargetScan software and the results
suggested that BDNF is a novel target gene of the
hsa-miR-183/96/182 cluster in RPE cells (Fig. 1B).
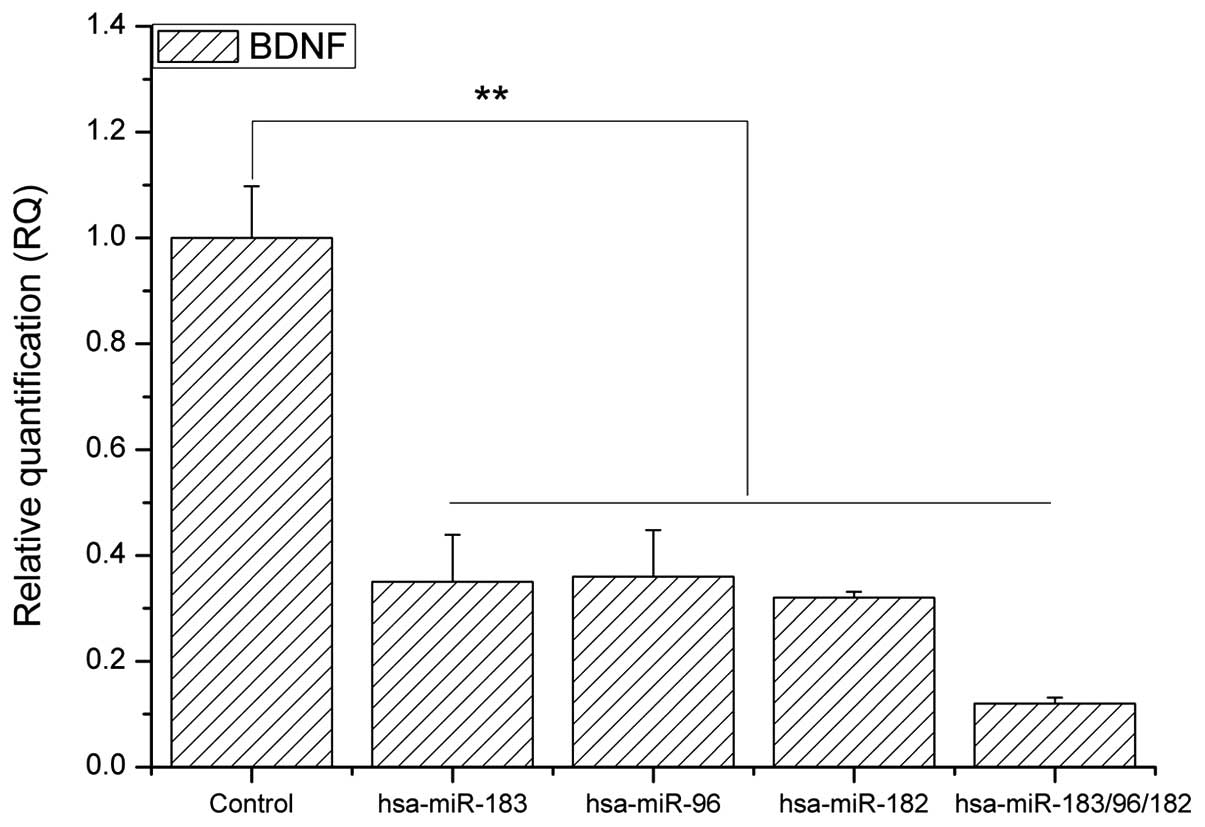
hsa-miR-183/96/182 mimics downregulates
the mRNA and protein expression of BDNF in RPE cells
In order to investigate the influence of the
hsa-miR-183/96/182 cluster on BDNF mRNA and protein expression in
RPE cells, mimic sequences of hsa-miR-183, hsa-miR-96, hsa-miR-182
and hsa-miR-183/96/182 were designed (Table I) The mimics were then transfected
into RPE cells. BDNF mRNA expression was significantly
downregulated in cells transfected with hsa-miR-183/96/182 mimics
when compared with BDNF mRNA expression in the control RPE cells,
including mimics of hsa-miR-183, hsa-miR-96, hsa-miR-182 (Fig. 2, P<0.01). Similarly, BDNF
protein expression was significantly downregulated in the RPE cells
transfected with hsa-miR-183/96/182 mimics, compared with that in
the control RPE cells (Fig. 3,
P<0.05; P<0.01).
 | Table ImiRNA mimic sequences. |
Table I
miRNA mimic sequences.
| Mimic name | Sequence
(5′→3′) |
|---|
| miR-182 | UUUGGCAAUGGUAGAACUCACACU |
| miR-183 | UAUGGCACUGGUAGAAUUCACU |
| miR-96 | UUUGGCACUAGCACAUUUUUGCU |
| miR-183/96/182 |
UUGGCAAAUGGCACUUGGCAC |
BDNF is a novel target gene of the
hsa-miR-183/96/182 cluster
In order to validate the targeting of the
hsa-miR-183/96/182 cluster to BDNF, a luciferase reporter vector
was designed (Fig. 4A). Following
transfection of hsa-miR-183/96/182 mimics into RPE cells, the ratio
of Renilla luciferase/firefly luciferase was found to be
significantly lower in mimic + wild type RPE cells, compared with
the ratios in the psiCHECK-2, wild type and mimic + mutant RPE
cells (Fig. 4B, P<0.01). The
ratio of Renilla luciferase/firefly luciferase in the RPE
cells of the wild type, mimic+mutant and psiCHECK-2 RPE cells were
not significantly different (Fig.
4B).
Discussion
The present study demonstrated that photostimulation
upregulated the expression of hsa-miR-183, hsa-miR-96 and
hsa-miR-182 in RPE cells. Artificially synthesized
hsa-miR-183/96/182 cluster mimics led to the downregulation of BDNF
mRNA and protein expression in RPE cells, and significantly
decreased the Renilla luciferase/firefly luciferase ratio in
the RPE cells of the mimic + wild type group. The results indicated
that BDNF is a novel target gene of the hsa-miR-183/96/182 cluster
and BDNF provides a novel target for the prevention and treatment
of light-induced retinal damage.
BDNF is an important member of the nerve growth
factor family (30,31). It targets certain neurons of the
central and peripheral nervous systems, and supports the survival
of existing neurons, thereby facilitating synapse growth and
differentiation (32–34). It is found in a number of tissues,
including the brain (35), retina
(36,37), central nervous system (38,39)
and motor neurons (40–42). In the retina, BDNF controls the
development of certain amacrine cell subtypes (43–45)
and regulates morphological maturation of ganglion cells (46–48).
Ladewig et al (49)
demonstrated that BDNF activates the N-methyl-D-aspartate receptor
to develop retinal ganglion cells. Rothe et al (50) showed that BDNF exhibits specific
ionic channels that are necessary for repetitive firing in early
retinal ganglion cell development. Numerous studies have documented
that BDNF is involved in increasing the survival of photoreceptors
in RPE cells that have undergone inherited retinal degeneration,
light damage and experimental retinal detachment (51,52).
Therefore, BDNF may be useful for the treatment and cure of
light-induced retinal damage. However, to the best of our
knowledge, there have been no studies investigating the regulation
of BDNF by miRNAs, such as miR-183/96/182.
The hsa-miR-183/96/182 cluster is a highly
conserved, intergenic sensory organ-specific and paralogous miRNA
cluster, and regulates an attractive candidate gene, as described
previously (26,53,54).
The hsa-miR-183/96/182 cluster is expressed in photoreceptor cells
and in the inner nuclear layer of the adult retina (24). The present study investigated the
hypothesis that the miR-183/96/182 cluster is involved in BDNF gene
regulation in the retina.
The expression of hsa-miR-183, hsa-miR-96 and
hsa-miR-182 was measured in RPE cells following exposure to visible
light or exposure to dark conditions. The results indicated that
hsa-miR-183, hsa-miR-96 and hsa-miR-182 expression in the RPE cells
is upregulated following exposure to visible light and
downregulated following maintenance in dark conditions.
Subsequently, hsa-miR-183/96/182 cluster mimics were synthesized
in vitro and their regulatory effect on BDNF expression was
investigated. According to a TargetScan software prediction, BDNF
is a novel target of hsa-miR-183/96/182 cluster mimics.
Furthermore, when RPE cells were transfected with
hsa-miR-183/96/182 mimics, the expression of BDNF mRNA and protein
was downregulated.
In conclusion, the results of the present study show
that BDNF is a novel target of the hsa-miR-183/96/182 cluster in
the retina. hsa-miR-183/96/182 mimics exhibited potential
protective mechanisms against light injury in RPE cells via the
BDNF gene. Therefore, the current study provides a novel approach
for the prevention and treatment of light-induced retinal
injury.
Acknowledgments
This study was supported by the China Postdoctoral
Science Foundation (grant no. 2013M532109).
References
|
1
|
Iandiev I, Wurm A, Hollborn M, et al:
Müller cell response to blue light injury of the rat retina. Invest
Ophthalmol Vis Sci. 49:3559–3567. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Vázquez-Chona F, Song BK and Geisert EE
Jr: Temporal changes in gene expression after injury in the rat
retina. Invest Ophthalmol Vis Sci. 45:2737–2746. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wang AG, Chen CH, Yang CW, et al: Change
of gene expression profiles in the retina following optic nerve
injury. Brain Res Mol Brain Res. 101:82–92. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Brod RD, Olsen KR, Ball SF and Packer AJ:
The site of operating microscope light-induced injury on the human
retina. Am J Ophthalmol. 107:390–397. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Henkes HE: Light injury to the retina.
Manifestation of acute posterior multifocal placoid pigment
epitheliopathy (author’s transl). Klin Monbl Augenheilkd.
170:813–818. 1977.In German. PubMed/NCBI
|
|
6
|
Meier-Koll A: Differential diagnosis of
injury to the retina by means of electrically evoked light
sensations (electrophosphene). Albrecht Von Graefes Arch Klin Exp
Ophthalmol. 184:177–192. 1972.In German. View Article : Google Scholar
|
|
7
|
Wilson KM, Lynch CM, Faraci FM and Lentz
SR: Effect of mechanical ventilation on carotid artery thrombosis
induced by photochemical injury in mice. J Thromb Haemost.
1:2669–2674. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Parver LM: Photochemical injury to the
foveomacula of the monkey eye following argon blue-green panretinal
photocoagu-lation. Trans Am Ophthalmol Soc. 98:365–374. 2000.
|
|
9
|
Costa E, Kharlamov A, Guidotti A, Hayes R
and Armstrong D: Sequelae of biochemical events following
photochemical injury of rat sensory-motor cortex: mechanism of
ganglioside protection. Patol Fiziol Eksp Ter. 4:17–23.
1992.PubMed/NCBI
|
|
10
|
Noell WK, Walker VS, Kang BS and Berman S:
Retinal damage by light in rats. Invest Ophthalmol. 5:450–473.
1966.PubMed/NCBI
|
|
11
|
Cai YS, Xu D and Mo X: Clinical,
pathological and photochemical studies of laser injury of the
retina. Health Phys. 56:643–646. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Rattner A, Yu H, Williams J, Smallwood PM
and Nathans J: Endothelin-2 signaling in the neural retina promotes
the endothelial tip cell state and inhibits angiogenesis. Proc Natl
Acad Sci USA. 110:E3830–E3839. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kawasaki H, Wadhwa R and Taira K: World of
small RNAs: from ribozymes to siRNA and miRNA. Differentiation.
72:58–64. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li CM, Zheng JG and Du GS: miRNA: a new
regulator of gene expression. Yi Chuan. 26:133–136. 2004.In
Chinese.
|
|
15
|
Luciano DJ, Mirsky H, Vendetti NJ and Maas
S: RNA editing of a miRNA precursor. RNA. 10:1174–1177. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Huang KM, Dentchev T and Stambolian D:
MiRNA expression in the eye. Mamm Genome. 19:510–516. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lee RC, Feinbaum RL and Ambros V: The C.
elegans heterochronic gene lin-4 encodes small RNAs with antisense
complementarity to lin-14. Cell. 75:843–854. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Reich J, Snee MJ and Macdonald PM:
miRNA-dependent translational repression in the Drosophila ovary.
PLoS One. 4:e46692009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lee YS, Nakahara K, Pham JW, et al:
Distinct roles for Drosophila Dicer-1 and Dicer-2 in the
siRNA/miRNA silencing pathways. Cell. 117:69–81. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Liang XH, Hart CE and Crooke ST:
Transfection of siRNAs can alter miRNA levels and trigger
non-specific protein degradation in mammalian cells. Biochim
Biophys Acta. 1829:455–468. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
McIver SC, Roman SD, Nixon B and
McLaughlin EA: miRNA and mammalian male germ cells. Hum Reprod
Update. 18:44–59. 2012. View Article : Google Scholar
|
|
22
|
Zhou P, Xu W, Peng X, et al: Large-scale
screens of miRNA-mRNA interactions unveiled that the 3′UTR of a
gene is targeted by multiple miRNAs. PLoS One. 8:e682042013.
View Article : Google Scholar
|
|
23
|
Fang L, Du WW, Yang X, et al: Versican
3′-untranslated region (3′-UTR) functions as a ceRNA in inducing
the development of hepatocellular carcinoma by regulating miRNA
activity. FASEB J. 27:907–919. 2013. View Article : Google Scholar
|
|
24
|
Zhu Q, Sun W, Okano K, et al: Sponge
transgenic mouse model reveals important roles for the microRNA-183
(miR-183)/96/182 cluster in postmitotic photoreceptors of the
retina. J Biol Chem. 286:31749–31760. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liu Y, Han Y, Zhang H, et al: Synthetic
miRNA-mowers targeting miR-183-96-182 cluster or miR-210 inhibit
growth and migration and induce apoptosis in bladder cancer cells.
PLoS One. 7:e522802012. View Article : Google Scholar
|
|
26
|
Tang H, Bian Y, Tu C, et al: The
miR-183/96/182 cluster regulates oxidative apoptosis and sensitizes
cells to chemotherapy in gliomas. Curr Cancer Drug Targets.
13:221–231. 2013. View Article : Google Scholar
|
|
27
|
Xu S, Witmer PD, Lumayag S, Kovacs B and
Valle D: MicroRNA (miRNA) transcriptome of mouse retina and
identification of a sensory organ-specific miRNA cluster. J Biol
Chem. 282:25053–25066. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Krol J, Busskamp V, Markiewicz I, et al:
Characterizing light-regulated retinal microRNAs reveals rapid
turnover as a common property of neuronal microRNAs. Cell.
141:618–631. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lumayag S, Haldin CE, Corbett NJ, et al:
Inactivation of the microRNA-183/96/182 cluster results in
syndromic retinal degeneration. Proc Natl Acad Sci USA.
110:E507–E516. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lüesse HG, Roskoden T, Linke R, Otten U,
Heese K and Schwegler H: Modulation of mRNA expression of the
neurotrophins of the nerve growth factor family and their receptors
in the septum and hippocampus of rats after transient postnatal
thyroxine treatment. I Expression of nerve growth factor,
brain-derived neurotrophic factor, neurotrophin-3 and neuro-trophin
4 mRNA. Exp Brain Res. 119:1–8. 1998. View Article : Google Scholar
|
|
31
|
Hohn A, Leibrock J, Bailey K and Barde YA:
Identification and characterization of a novel member of the nerve
growth factor/brain-derived neurotrophic factor family. Nature.
344:339–341. 1990. View
Article : Google Scholar : PubMed/NCBI
|
|
32
|
Alvarez-Borda B, Haripal B and Nottebohm
F: Timing of brain-derived neurotrophic factor exposure affects
life expectancy of new neurons. Proc Natl Acad Sci USA.
101:3957–3961. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Chmielnicki E, Benraiss A, Economides AN
and Goldman SA: Adenovirally expressed noggin and brain-derived
neurotrophic factor cooperate to induce new medium spiny neurons
from resident progenitor cells in the adult striatal ventricular
zone. J Neurosci. 24:2133–2142. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Pencea V, Bingaman KD, Wiegand SJ and
Luskin MB: Infusion of brain-derived neurotrophic factor into the
lateral ventricle of the adult rat leads to new neurons in the
parenchyma of the striatum, septum, thalamus and hypothalamus. J
Neurosci. 21:6706–6717. 2001.PubMed/NCBI
|
|
35
|
von Diemen L, Kapczinski F, Sordi AO, et
al: Increase in brain-derived neurotrophic factor expression in
early crack cocaine withdrawal. Int J Neuropsychopharmacol.
17:33–40. 2014. View Article : Google Scholar
|
|
36
|
Abu El-Asrar AM, Nawaz MI, Siddiquei MM,
Al-Kharashi AS, Kangave D and Mohammad G: High-mobility group box-1
induces decreased brain-derived neurotrophic factor-mediated
neuroprotection in the diabetic retina. Mediators Inflamm.
2013:8630362013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Chen R, Yin XB, Peng CX and Li GL: Effect
of brain-derived neurotrophic factor on c-jun expression in the rd
mouse retina. Int J Ophthalmol. 5:266–271. 2012.PubMed/NCBI
|
|
38
|
Numakawa T, Adachi N, Richards M, Chiba S
and Kunugi H: Brain-derived neurotrophic factor and
glucocorticoids: reciprocal influence on the central nervous
system. Neuroscience. 239:157–172. 2013. View Article : Google Scholar
|
|
39
|
Yan Q, Rosenfeld RD, Matheson CR, et al:
Expression of brain-derived neurotrophic factor protein in the
adult rat central nervous system. Neuroscience. 78:431–448. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Niu C and Yip HK: Neuroprotective
signaling mechanisms of telomerase are regulated by brain-derived
neurotrophic factor in rat spinal cord motor neurons. J Neuropathol
Exp Neurol. 70:634–652. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Fryer HJ, Wolf DH, Knox RJ, et al:
Brain-derived neurotrophic factor induces excitotoxic sensitivity
in cultured embryonic rat spinal motor neurons through activation
of the phosphati-dylinositol 3-kinase pathway. J Neurochem.
74:582–595. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Wang W, Salvaterra PM, Loera S and Chiu
AY: Brain-derived neurotrophic factor spares choline
acetyltransferase mRNA following axotomy of motor neurons in vivo.
J Neurosci Res. 47:134–143. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Fujieda H and Sasaki H: Expression of
brain-derived neurotrophic factor in cholinergic and dopaminergic
amacrine cells in the rat retina and the effects of constant light
rearing. Exp Eye Res. 86:335–343. 2008. View Article : Google Scholar
|
|
44
|
Cellerino A, Arango-González BA and Kohler
K: Effects of brain-derived neurotrophic factor on the development
of NADPH-diaphorase/nitric oxide synthase-positive amacrine cells
in the rodent retina. Eur J Neurosci. 11:2824–2834. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Cellerino A and Kohler K: Brain-derived
neurotrophic factor/neurotrophin-4 receptor TrkB is localized on
ganglion cells and dopaminergic amacrine cells in the vertebrate
retina. J Comp Neurol. 386:149–160. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Leake PA, Hradek GT, Hetherington AM and
Stakhovskaya O: Brain-derived neurotrophic factor promotes cochlear
spiral ganglion cell survival and function in deafened, developing
cats. J Comp Neurol. 519:1526–1545. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Bonnet D, Garcia M, Vecino E, Lorentz JG,
Sahel J and Hicks D: Brain-derived neurotrophic factor signalling
in adult pig retinal ganglion cell neurite regeneration in vitro.
Brain Res. 1007:142–151. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Nakazawa T, Tamai M and Mori N:
Brain-derived neurotrophic factor prevents axotomized retinal
ganglion cell death through MAPK and PI3K signaling pathways.
Invest Ophthalmol Vis Sci. 43:3319–3326. 2002.PubMed/NCBI
|
|
49
|
Ladewig T, Fellner S, Zrenner E, Kohler K
and Guenther E: BDNF regulates NMDA receptor activity in developing
retinal ganglion cells. Neuroreport. 15:2495–2499. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Rothe T, Bähring R, Carroll P and Grantyn
R: Repetitive firing deficits and reduced sodium current density in
retinal ganglion cells developing in the absence of BDNF. J
Neurobiol. 40:407–419. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Zhang M, Mo X, Fang Y, et al: Rescue of
photoreceptors by BDNF gene transfer using in vivo electroporation
in the RCS rat of retinitis pigmentosa. Curr Eye Res. 34:791–799.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Caffé AR, Söderpalm AK, Holmqvist I and
van Veen T: A combination of CNTF and BDNF rescues rd
photoreceptors but changes rod differentiation in the presence of
RPE in retinal explants. Invest Ophthalmol Vis Sci. 42:275–282.
2001.PubMed/NCBI
|
|
53
|
Weeraratne SD, Amani V, Teider N, et al:
Pleiotropic effects of miR-183~96~182 converge to regulate cell
survival, proliferation and migration in medulloblastoma. Acta
Neuropathol. 123:539–552. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Mihelich BL, Khramtsova EA, Arva N, et al:
miR-183-96-182 cluster is overexpressed in prostate tissue and
regulates zinc homeostasis in prostate cells. J Biol Chem.
286:44503–44511. 2011. View Article : Google Scholar : PubMed/NCBI
|