Introduction
Prostate cancer refers to epitheliogenic malignant
tumors of the prostate. What is usually termed prostate cancer
refers to adenocarcinoma of the prostate. Since the mid to late
1980s, the morbidity and mortality rates of prostate cancer in
certain developed areas in China have continued to increase
(1). Previous studies have
indicated that seminal plasma protein is one of the specific
markers of prostate cancer (2).
Hao and Liang (3) used 131I to
mark the anti-human seminal plasma protein single-chain antibody
(γ-Sm-McAb), which was used for the radioimmunoimaging and
treatment of prostate cancer. This method was confirmed to be of
high sensitivity and specificity, and of beneficial therapeutic
effect (3). However, the
monoclonal antibodies used originated from murine animals, the
repeated use of which may generate anti-mouse antibodies, affecting
the therapeutic effect and resulting in allergic reactions
(4). Due to the small molecular
size, low antigenicity and advantages over other parental
antibodies when used within the body, a single chain variable
fragment (ScFv) can be used to effectively avoid deficiencies in
monoclonal antibodies and has received increasing attention
(5). A ScFv is composed of a heavy
chain (VH) and a light chain (VL) through linkers of several amino
acids (6) (Fig. 1). VH and VL are the minimum
functional fragments in the binding site of antibodies to antigens,
while the linker peptide is used predominantly for connection
between the two for fusion expression. (Gly4Ser)n is usually
selected as the linker peptide in ScFv (7), however, the effect of the of the
selected n-value on the functional expression of the two variable
regions in ScFv is key to investigating the optimal construct of
ScFv. In the present study, a similarity algorithm of spherical
coordinates for layered proteins was used. Comparison of changes in
the structural models of the changeable structures in single-chain
antibodies under different values of n, enable the optimal linker
length to be determined for maintaining active seminal plasma
protein single-antibodies. The results of the present study aim to
provide a foundation for the preparation of a single-chain
antibody, which may be effective in the treatment of prostate
cancer.
Materials and methods
Preparation of monoclonal antibody
The anti-human seminal plasma protein hybridoma cell
strain was provided by The First Affiliated Hospital of Zhengzhou
University (Zhengzhou, China), which was used for secretion of the
monoclonal antibody of anti-human seminal plasma protein. Plasmids
of Escherichia coli JM109 (pUC19) were preserved in the
laboratory (First Affiliated Hospital of Zhengzhou University). The
restriction endonuclease, Taq DNA polymerase, dNTP, buffer,
purification system of Wizard™Plus Minipreps and the expression
vector of the Glutathione S-transferase fusion protein (pGEX-4T-1)
were purchased from Promega Corporation (Fitchburg, WI, USA); The
rapid DNA ligation kit was purchased from Boehringer Mannheim GmbH
(Basel, Switzerland); The Advantage™PCR-Pure kit was purchased from
Clontech Co. (Tokyo, Japan).
The key instruments used in the present study
included a polymerase chain reaction (PCR) DNA Amplifier, a
pipettor and refrigerated centrifuge (Eppendorf, Hamburg, Germany),
a GFL-7601 incubator (GFL Company, Lower Saxony, Germany), a water
bath (Shanghai Senxin Biological Technology Co., Ltd., Shanghai,
China), a horizontal electrophoresis apparatus, a gel imaging
system (Liyyi Company, Beijing, China) and a −80°C ultra cold
storage freezer (Haier, Beijing, China).
Extraction and reverse transcription of
total cellular (c)DNA
The extraction of total cDNA was performed was
performed using the DNA one step extraction method, as described
previously (8). The total RNA was
extracted from the cells and dissolved in 30 µl RNase-free
water, and reverse transcription was performed using 5 µl,
according to the manufacturer's instructions (Promega
Corporation).
Amplification of variable region genes of
the monoclonal antibody
The genes of VH and VL were amplified using 672
ng/µl template cDNA. The primer sequences were as follows:
VH, reverse 5′-AGGT (CG) (AC) A (AG) CTGCAG (CG) AGTC (AT) GG-3′
(degenerate primer) and forward
5′-TGAGGAAACGGTGACCGTGGTCCCTTGGCCCCAG-3′; VL, reverse
5′-GTGAATTCGACATCGTGATGACCCAGTCTCC-3′ and forward
5′-CAGTCGACTAACGTTTGATCTCCAGCTTGGTCCC-3′ (Sangon Biotech, Shanghai,
China). The PCR reaction system (20 µl) included cDNA (1
µl), dNTP (0.4 µl), upstream and downstream primers
(each 0.3 µl), Taq DNA polymerase (0.2 µl), buffer (2
µl) and sterilized water (15.8 µl). The reaction
conditions included a denaturing stage for 60 sec at 94°C,
annealing for 90 sec at 55°C, extension for 120 sec at 72°C, for 30
cycles. Following extracting 5 µl amplicon from each of the
VH and VL samples to perform the lipid sugar electrophoresis
detection, the remaining amplicon was recycled by
Advantage™PCR-Pure kit according to the manufacturer's instructions
and then was sequenced by the Shenggong Bioengineering Limited
Company (Shanghai, China).
Modeling
The corresponding amino acid sequence was acquired
by translating VH and VL gene sequences in γ-Sm-ScFv. The amino
acid sequence of the protein determined the advanced structure.
Firstly, tertiary-structure modeling of VH and VL were performed
using the homologous modeling method. The amino acid sequences of
VH and VL were sent to SWISS-MODEL for modeling (http://www.swissmodel.expasy.org/), and the
acquired data file of the protein data bank structure was treated
as the original structural file.
A total ScFv sequence can be combined by adding
linker amino acid sequences and connecting them between VH and VL.
Among which, (Gly4Ser)n was selected for the linker sequence.
(Gly4Ser) refers to five short amino acid peptides, which are
composed of four glycines and one serine. This oligopeptide is a
repeated unit in the linker peptide, which is used for ligation of
the VL and VH segments. It has relatively high flexibility or
bending capacity. The small steric hindrance is useful for the
interaction of the VL and VH segments, and arrangement of the
correct conformation, as well as improving the stability of
antibodies. Such structure is not easily recognized or degraded by
proteases, which contributes to the stability of antibodies within
the body. The combinatorial ScFv-n sequence was also used in the
modeling, and the data of the overall three-dimensional structure
and that of VHn and VLn were obtained.
Structural contrast
To investigate the effect of the length of the
linker peptide on the structure of VH and VL, the structural
similarity of VH and VL in (Gly4Ser)n was calculated at different
values of n. In addition, a space spherical shell hierarchical
matching algorithm, based on spherical coordinates and described by
Zhang and Chen (9), was used to
calculate the level of similarity. In this algorithm, the protein
was treated as the spherome, and the euclidean coordinates of each
atom in the protein were transformed into spherical coordinates.
According to the radius, the protein was then divided into several
layers of spherical shells, and the same number of same atoms in
each layer were collected. On the basis of predetermined weights,
the number of atoms were determined, thus, obtaining the final
value of atoms in each layer. Each layer of atomic values of
VH-(Gly4Ser)n or VL-(Gly4Ser)n and VH (or VL) were stacked into
vector a and vector b, respectively. The included angle cosine
function was treated as a similarity function. The formula used
(1), the algorithm of which was
achieved using MATLAB 2012a software (The MathWorks, Inc., Natick,
MA, USA) was as follows:
Stability analysis
The stability of ScFv is presented as the fore and
aft distance or the diffusion radius. The fore and aft distance
refers to the distance from the fore α-carbon atom to the aft
α-carbon atom. Using MATLAB, the fore and aft distances of the
original structure of VH and VL were calculated, respectively,
adding the fore and aft distances of VHn and VLn when n=0–9. In
addition, their largest diffusion radius was calculated.
Results
The optical density
(OD)260/OD280 value of the overall extracted
mRNA was 2.1, according and its concentration was 1,279
ng/µl, which met the requirements of the experiment. The
concentration of cDNA transcribed by reverse transcription was 672
ng/µl. The cDNA was diluted at 1:50 and was used to amplify
of the VH and the VL of the seminal plasma protein's single-chain
antibody using PCR. The results of the electrophoresis are shown in
Fig. 2.
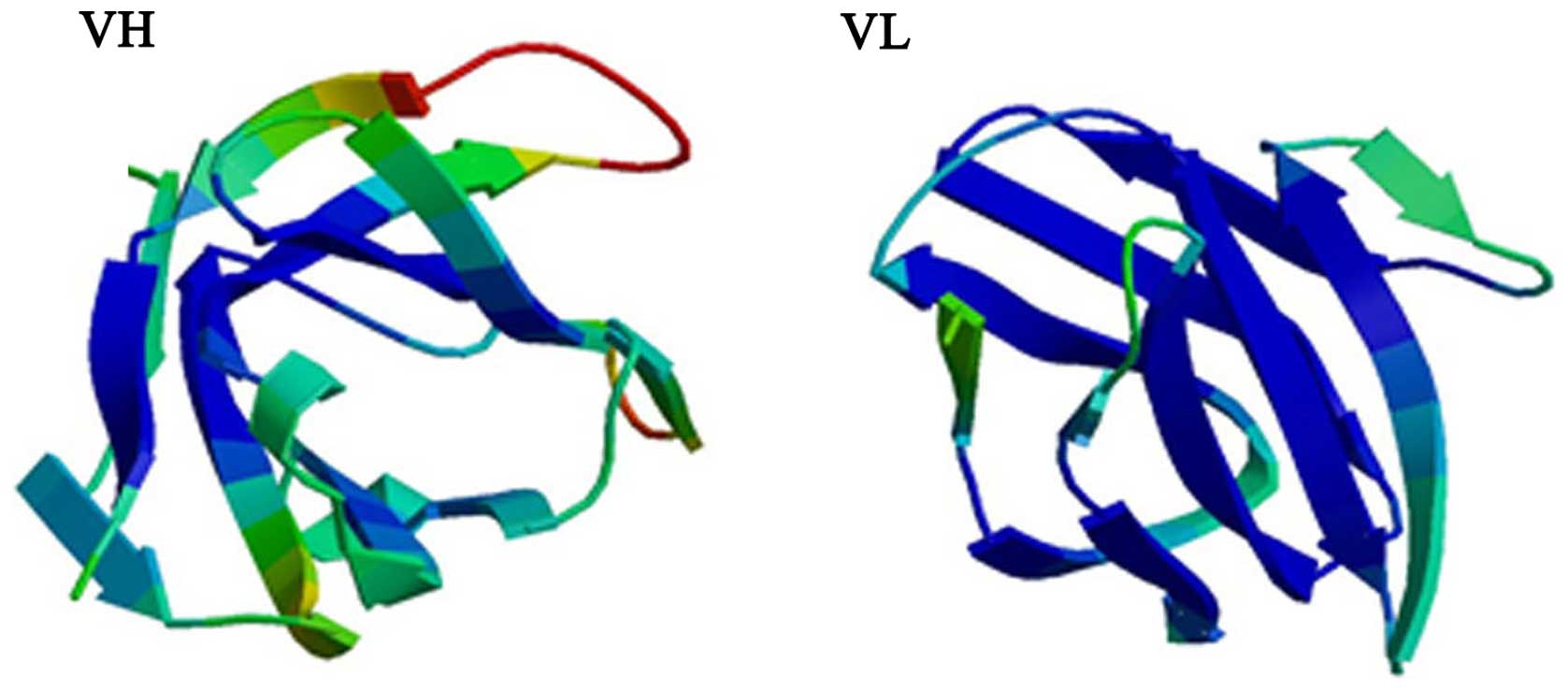
The results of the VH and VL modeling in ScFv using
the homologous modeling method are shown in Figs. 3 and 4. Generally, the length of the linker
peptide of ScFv was between 10 and 40 amino acids long. The longer
the length of the linker peptide, the less stable the structure of
the ScFv. During the process of modeling, when n=9 (linker peptide
length of 45 amino acids), modeling failed due to the low homology
of the searched templates. Therefore, three-dimensional models were
obtained when the n-value was between 0 and 8, and the results of
the modeling are shown in Fig. 4.
Each model had corresponding data for the VHn and VLn structure and
data for the overall ScFv-n structure.
From the figures described above, there was a clear
difference in the structural models between VH/VL alone and VH/VL
when the linker was added, and the structure of the linker changed
markedly as its length changed. Subsequently, the protein
similarity algorithm, based on delamination of the spherical
coordinates, was used to provide a contrast in the VH and VL
similarity prior to and following addition of the linker. The
results of the algorithm are shown in Table I.
 | Table IResults of similarity using a space
spherical shell hierarchical matching algorithm. |
Table I
Results of similarity using a space
spherical shell hierarchical matching algorithm.
| Chain | Similarity
| Average |
|---|
| Layer 2 | Layer 3 | Layer 4 |
|---|
| VH0 | 0.8910 | 0.7653 | 0.6183 | 0.7582 |
| VH1 | 0.9033 | 0.6320 | 0.7680 | 0.7678 |
| VH2 | 0.9029 | 0.7931 | 0.5616 | 0.7525 |
| VH3 | 0.9076 | 0.8376 | 0.5866 | 0.7773 |
| VH4 | 0.9073 | 0.8198 | 0.5866 | 0.7712 |
| VH5 | 0.9083 | 0.8299 | 0.5825 | 0.7736 |
| VH6 | 0.9085 | 0.8299 | 0.5825 | 0.7736 |
| VH7 | 0.9085 | 0.8258 | 0.5861 | 0.7735 |
| VH8 | 0.9085 | 0.8258 | 0.5861 | 0.7735 |
| VL0 | 0.9482 | 0.9723 | 0.8021 | 0.9075 |
| VL1 | 0.9515 | 0.9726 | 0.7992 | 0.9078 |
| VL2 | 0.9515 | 0.9729 | 0.7992 | 0.9079 |
| VL3 | 0.9515 | 0.9734 | 0.7997 | 0.9082 |
| VL4 | 0.9515 | 0.9726 | 0.7992 | 0.9078 |
| VL5 | 0.9515 | 0.9729 | 0.7992 | 0.9079 |
| VL6 | 0.9515 | 0.9734 | 0.8018 | 0.9089 |
| VL7 | 0.9507 | 0.9734 | 0.7997 | 0.9079 |
| VL8 | 0.9507 | 0.9734 | 0.7997 | 0.9079 |
Following several trials, the similarity data was
relatively accurate and stable when the protein was divided into
three layers. To guarantee the accuracy of the results, the protein
was divided into 2, 3 and 4 layers. Subsequently, the mean
similarity value of the data in each group was calculated and
treated as the final result. As shown in Table I, when n=3 and n=6, the similarity
data obtained was close to the original data of VH and VL. The
similarity has indicated the effect of the linker peptide's length
on the variable region structure to the ScFv, and the larger the
value of the similarity, the lower the effect, which represented
that the effect on the biological activity of a single-chain may be
smaller.
Following the addition of the linker, the stability
of VH and VL was analyzed again, based on the fore and aft distance
and the diffusion radius. The results are shown in Table II.
 | Table IIChanges of the fore and aft distance
and radius of VH, VL and linker prior to and following addition of
the linker. |
Table II
Changes of the fore and aft distance
and radius of VH, VL and linker prior to and following addition of
the linker.
| Chain | Fore and aft
distance | Radius | Length of linker | Fore and aft distance
of linker |
|---|
| VH0 | 32.9989 | 23.2127 | | |
| VH1 | 42.3431 | 35.2561 | | |
| VH2 | 37.1628 | 26.1484 | | |
| VH3 | 41.6467 | 25.6413 | | |
| VH4 | 38.8902 | 27.3090 | | |
| VH5 | 40.5697 | 28.6009 | | |
| VH6 | 40.3791 | 28.3365 | | |
| VH7 | 40.8331 | 28.1269 | | |
| VH8 | 40.8454 | 28.1225 | | |
| VH | 39.9601 | 25.2179 | | |
| VL0 | 22.3826 | 25.8404 | 0 | – |
| VL1 | 19.0852 | 25.8461 | 5 | 14.021 |
| VL2 | 19.0833 | 25.8452 | 10 | 13.5976 |
| VL3 | 19.0739 | 25.8434 | 15 | 17.3828 |
| VL4 | 19.0852 | 25.846 | 20 | 25.2548 |
| VL5 | 19.0740 | 25.8428 | 25 | 16.7704 |
| VL6 | 19.0659 | 25.8402 | 30 | 16.5636 |
| VL7 | 19.0846 | 25.8444 | 35 | 17.7174 |
| VL8 | 19.0745 | 25.8442 | 40 | 18.0869 |
| VL | 36.7271 | 22.6299 | – | – |
As shown in Table
II, when n=6, the fore and aft distance (40.3791) of the VH was
the closest to that of VH (39.9601); when n=3, in which the
diffusion radius (25.6413) was also closest to the original data
(25.2179). By contrast, in the VL, the fore and aft distance
following addition of the linker reduced to almost half that of the
original data, however, the diffusion radius increased. In
addition, when n≤3, the fore and aft distance of the linker
increased as the length of the peptide increased, whereas, when
n>3, the fore and aft distance decreased due to folding of the
lengthened peptide chain. The changes in the fore and aft distance
also affected VH and VL. The position comparison in the space
coordinates system of the original VH, VL and the α carbon atoms of
the ScFv-n structure was established by MATLAB 2012a, as is shown
in Figs. 5 and 6, which suggested the fore and aft
distance change of VH and VL was as expected. This indicated that
after adding linker peptides of different lengths, the change in
the whole molecular radius was small.
Discussion
From the comparative investigation performed in the
present study, it was concluded that, in the spatial distribution
of analogous proteins, the three-dimensional structure of proteins
was more conserved than the primary sequence. The prediction of
target protein spatial conformation is more reliable when performed
in analogous proteins (10), and
there have been several previous successes in predicting the
structure of antibodies (10,11).
Successful modeling of the 6B4 antithrombotic antibody by Fontayne
et al (11) laid a
foundation for the investigation of its antithrombotic activity and
antigenicity Modeling of the three-dimensional structure of a
protein can assist in the modification of the original antibodies
on the basis of understanding its spatial structure and
physiological functions, and it is significant for antibody
engineering. ScFv molecule is a small molecule with weak
immunogenicity and a rapid response time. It has no cumulative
action in the kidney and its sharpness and clarity in tumor imaging
is high, which enable it to be used as a vector and combined with
medicine, isotopes and toxins (12). Single-chain antibody constructs
belong to minimolecular antibodies. They are constructed by VH, VL
and linker peptides, and are the smallest functional fragments of
antibodies in retention of the antigen-binding site, which has
significant theoretical and applicable value in the diagnosis and
treatment of cancer.
In the present study, a series of methods, including
modeling and bioinformatics, were used to examine and compare the
structural changes in ScFv when the length of the linker peptide
was altered. (Gly4Ser)n, as a linker peptide, is often selected in
single-chain antibodies, among which 'Gly' represents glycine,
'Ser' represents serine and 'n' indicates the number of (Gly4Ser).
The results of the present study revealed that, following addition
of the linker, the similarity data for VH and VL was closest to
that of the original data when n=3 and n=6, which indicated that
ScFv-3 and ScFv-6 were closest to the original structure, as was
their biological activity. In terms of stability, the fore and aft
distance, and the diffusion radius of the VH and VL altered to
different degrees following addition of the linker. Notably, when
n=6, the fore and aft distance of the VH was closest to the
original data, while that of the VL decreased by half. Compared
with the original radius, that of VH and VL increased. When n=3,
however, the diffusion radius of the VH was the closest to the
original data. The majority of the results indicated the the
significance of the effect of linkers n=3 and n=6 on VH and VL.
Despite the construction of ScFv or a single-strand
bispecific antibody, the linker peptide cannot affect the folding
of the structure of an antibody or its biological activities
(13). Several studies have
investigated the effect of the length of linkers on the activities
of ScFv (14). Kikuchi et
al (15) constructed an
anti-CD47 bivalent single-chain antibody, MABLsc(Fv), by way of
covalence, using linkers of 15 amino acids (Gly4Ser); Yan et
al (16) found that a linker
length of 15 amino acids was more favorable for the folding of
antibodies, guaranteeing the affinity of the bivalent single-chain
antibodies. Goel et al (17) successfully constructed bivalent
single-chain antibodies with immune activity using a longer linker
of 25 amino acids. The present study confirmed that when n=3
(linker with 25 amino acids), the two variable regions in the ScFv
were closest to the original structure. The results also
demonstrated that, when n=6, the effect of the linker on the
structure of ScFv was smaller. Considering that the present study
used a bioinformatics approach, and that there may be a certain
level of error in molecular modeling, the results of the present
study require confirmation using biological experiments. Whether a
linker length of 30 amino acids is suitable for the favorable
expression of biological activity in a single-chain antibody also
requires further investigation.
The effects of structural changes of linker peptides
in single-chain antibodies on the whole antibody molecules were
examined using mathematical modeling and bioinformatics methods,
providing a basis for further investigation of the preparation of
single-chain antibodies.
Acknowledgments
The authors would like to thank their colleagues
within the department for their general support. This study was
supported by the National Natural Science Foundation of China
(grant no. 60971110), the Science and Technology Corporation
Project of Henan Province (grant no. 122106000042) and the Open
Science and Technology Corporation Project of Henan Province in
2013 (grant no. 132106000064).
References
|
1
|
Peng P, Gong YM, Bao PP, et al: Estimates
and prediction of prostate cancer incidence, mortality and
prevalence in China in 2008. Zhonghua Liu Xing Bing Xue Za Zhi.
33:1056–1059. 2012.In Chinese.
|
|
2
|
Arai Y, Yoshiki T, Okada K and Yoshida O:
Multiple marker evaluation in prostatic cancer using prostatic
specific antigen, gamma-seminoprotein and prostatic acid
phosphatase. Urol Int. 44:135–139. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hao XK and Liang GD: Diagnosis of prostate
cancer by radioim-munoimaging of 131I-antihuman seminal plasma
protein. Natl Med J China. 73:655–657. 1993.
|
|
4
|
Hao XK, Wu GJ, Bai YJ, Yao LB, Zhang YH
and Gao L: Construction and expression of single-chain Fv Genes of
Anti-human seminal plasma protein. Chin J Urol. 29:30–32. 1999.
|
|
5
|
Fei JX, Ren J and Miao JY: Research on the
Construction, expression and clinical application of single-chain
antibody. Shaanxi Med J. 29:30–32. 2000.
|
|
6
|
Wu HG and Yan DF: Construction and
expression of the variable region of single-chain antibody. J
Taizhou Teach Coll. 17:55–59. 1995.
|
|
7
|
Yang LJ, Hou YC, Bai YJ, Yao LB and Su CZ:
Analysis of primary structure and modeling of spatial structure of
single-chain variable region of antibody against human gastric
cancer. Shijie Huaren Xiaohua Zazhi. 16:2333–2336. 2008.
|
|
8
|
Chomczynski P and Sacchi N: Single-step
method of RNA isolation by acid guanidinium
thiocyanate-pheno-chloro form extraction. Anal Biochem.
162:156–159. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang JH and Chen ZT: Analyzing influence
on the conformation of single-chain antibody with the differential
length of linkers. Afr J Microbiol Res. 5:5737–5744. 2011.
|
|
10
|
Thompson AJ, Price KL, Reeves DC, Chan SL,
Chau PL and Lummis SC: Locating an antagonist in the 5-HT3 receptor
binding site using modeling and radioligand binding. J Biol Chem.
280:20476–20482. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Fontayne A, Vanhoorelbeke K, Pareyn I, Van
Rompaey I, Meiring M, Lamprecht S, Roodt J, Desmet J and Deckmyn H:
Rational humanization of the powerful antithrombotic anti-GPIbalpha
antibody: 6B4. Thromb Haemost. 96:671–684. 2006.PubMed/NCBI
|
|
12
|
Colcher D, Bird R, Roselli M, Hardman KD,
Johnson S, Pope S, Dodd SW, Pantoliano MW, Milenic DE and Schlom J:
In vivo tumor targeting of a recombinant single-chain
antigen-binding protein. J Natl Cancer Inst. 82:1191–1197. 1990.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Fang Ming, Jiang Xin and Yang Zhi:
Influences of connecting peptide between the chain on the
biological activity of ScBsAb. Chin Sci Bull. 48:1912–1918.
2003.
|
|
14
|
Zhang J, Shang Z and Zhang X: The
optimization design of single bivalent connecting peptide
antibodies. Math Med Biol. 21:602–605. 2008.
|
|
15
|
Kikuchi Y, Suzuki Y and Tamiya N: The
source of oxygen in the reaction catalysed by collagen lysyl
hydroxylase. Biochen J. 213:507–512. 1983.
|
|
16
|
Yan D, Fang J and Song J: Construction and
Expression of Bivalent Single-chain Antibodies with Different
Linker Sequence Against Human Colorectal Carcinoma. J Cell Biol.
29:272–276. 2007.
|
|
17
|
Goel A, Colcher D, Baranowska-Kortylewicz
J, et al: Genetically engineered tetravalent single-chain Fv of the
pan carcinoma monoclonal antibody CC49: improved bio distribution
and potential for therapeutic application. Cancer Res.
60:6964–6971. 2000.
|