Introduction
Liver cancer is one of the most lethal malignancies
with a clinically poor prognosis. There are two main reasons for
this poor prognosis: One is the difficulty in the early detection
of liver cancer. Patients with clinical symptoms are already at an
advanced cancer stage, meaning that they have already missed the
chance for surgical treatment. The second is the current lack of
effective chemotherapeutic treatment for patients with liver
cancer. No drug treatments are currently available for patients
with liver cancer (1). In recent
years, with a greater understanding of the molecular mechanisms of
cancer, targeted therapy has become a novel concept for tumor
treatment. Targeted therapy achieves its therapeutic purposes by
means of targeting those molecules, which are important in tumor
growth, invasion or metastasis. The majority of common targets are
the critical molecules in cell signaling pathways, and interfering
with these molecules may block the respective signaling pathway and
lead to tumor growth inhibition or apoptosis (2).
The association between Ras homolog gene family,
member C (RhoC) overexpression and tumor development has become an
intense area of interest in recent years. A growing number of
investigations have utilized RhoC as a target molecule for cancer
gene therapy (3–5). RhoC has a marked impact on local
tumor cell invasion and distant metastasis through regulation of
the actin cytoskeleton. Its elevated expression in hepatoma cells
promotes tumor invasion and distant metastasis (6). In addition, RhoC overexpression is
also an important factor in promoting hepatoma cell proliferation
(7). RhoC also induces malignant
transformation of normal liver cells (8). Therefore, RhoC overexpression is
essential for the survival, invasion and metastasis of
hepatocellular carcinoma cells. Overexpression of RhoC is also
important in the invasion and distant metastasis of liver cancer
cells. RhoC as a molecular target is expected to produce good
results for hepatocellular carcinoma treatment.
However, the mechanisms of tumor cell survival and
invasion are complex, and specific control of the growth and
metastasis of tumor cells is difficult. Therefore, researchers have
begun to adopt a joint approach to eradicate or inhibit the growth
of tumor cells. The phosphoinositide 3 kinase (PI3K)/Akt/mammalian
target of rapamycin (mTOR) signal transduction pathway is closely
associated with tumor cell proliferation, invasion and apoptosis,
and has thus become an area of interest in the study of liver
cancer (9). Rapamycin (RAPA) is a
well-known specific inhibitor of the PI3K/Akt/mTOR signaling
pathway. Thus, inhibition of mTOR activity may lead to tumor cell
growth suppression.
RAPA does not act directly on the mTOR molecule. It
first binds with the FK506 protein to form a complex. This complex
attaches with the FRB domain of mTOR, disturbing mTOR signaling,
affecting the synthesis of associated proteins, and therefore
inhibiting mTOR activity (10).
RAPA has potent clinical effects against cervical cancer,
endometrial cancer and ovarian cancer (11). Its derivatives, including letrozole
and everolimus, are able to prohibit the growth and progression of
breast cancer, kidney cancer and pancreatic cancer, respectively
(11–13). For patients with liver cancer, RAPA
and its derivatives may also have a good therapeutic prospect. In
addition to the direct inhibition of tumor cell proliferation, RAPA
also inhibits the proliferation of vascular endothelial growth
factor (VEGF) and vascular endothelial cells, which are important
in cancer therapy (14,15). It is known that the RhoC and
PI3K/Akt/mTOR pathways are closely linked (13). RhoC overexpression leads to
PI3K/Akt/mTOR pathway activation in melanoma cells, thus promoting
melanoma metastasis (16).
Following PI3K/Akt/mTOR pathway activation, the activated Akt
molecule phosphorylates RhoC, promoting breast cancer cell
metastasis (17). Thus, the RhoC
and the PI3K/Akt/mTOR pathway are able to activate each other and
are important in tumor growth. Therefore, it was hypothesized that
the joint effort of targeting RhoC and the PI3K/Akt/mTOR pathway
may produce synergistic effects in liver cancer treatment.
Therefore, the present study selected hepatocellular carcinoma
cells to investigate the combined effects of RhoC gene silencing
and the PI3K/Akt/mTOR pathway-specific inhibitor RAPA on the
biological behavior of hepatocellular carcinoma cells, including
proliferation, invasion and migration, providing theoretical and
experimental evidence for liver cancer treatment.
Materials and methods
Reagents and antibodies
pU6mRFP RhoC-small interfering (si)RNA and pU6mRFP
scramble-small interfering (si)RNA plasmids were constructed in our
laboratory (Department of General Surgery, First Hospital of Jilin
University, Changchun, China). The DNA molecular weight standard
DL2000 and the reverse transcriptional polymerase chain reaction
(RT-PCR) kit (RNA PCR kit version 3.0) were purchased from Takara
Biotechnology Co., Ltd. (Dalian, China). SuperFect Transfection
Reagent (cat. no. 301305) was purchased from Qiagen China Co., Ltd.
(Shanghai, China). RAPA was purchased from LC Laboratories (Woburn,
MA, USA). The primary mouse anti-human β-actin antibody (cat. no.
MA5-15739; 1:500) was purchased from Invitrogen Life Technologies
(Carlsbad, CA, USA), goat anti-mouse secondary antibody (cat. no.
sc-2005; 1:500), goat anti-rabbit secondary antibody (cat. no.
sc-2030; 1:500) and rabbit anti-human RhoC polyclonal antibody
(cat. no. sc-28565; 1:200).
Cell culture and transfection
BEL7402 human hepatoma cells were purchased from
Shanghai Institute of Biochemistry and Cell Biology Cell Bank
(Shanghai, China). The cells were cultured at 37°C in
4-(2-hydroxyethyl)-1-piperazineethane-sulfonic acid-buffered
Dulbecco's modified Eagle's medium (H-DMEM; Invitrogen Life
Technologies) in a 5% CO2 incubator (Shanghai Shen-Li
High Tech Co. Ltd., Shanghai, China). Cells in the logarithmic
growth phase were seeded into 24-well plates until the cell density
reached ~70% confluence. Transfection was performed according to
the manufacturer's instructions. Cells were cultured in an
incubator following transfection, and the culture medium was
discarded 4 h later. Cells were washed with phosphate-buffered
saline (PBS) solution, and 400 µl fresh H-DMEM containing
10% fetal bovine serum, 1% penicillin and 1% streptomycin (all
Invitrogen Life Technologies) was added to each well.
BEL7402 cells were used as the blank control group,
BEL7402 cells transfected with pU6mRFP scramble-siRNA were used as
the negative control group, and BEL7402 cells transfected with
pU6mRFP RhoC-siRNA were used as the RNA interference (RNAi) group.
Following 72 h of incubation, 1×106 cells/ml were
harvested and lysed with TRIzol reagent (Invitrogen Life
Technologies) for total RNA extraction. RNA was then reverse
transcribed to cDNA, according to the manufacturer's instructions.
Semi-quantitative RT-PCR was performed to detect RhoC mRNA
expression levels. PCR was conducted using the RNA PCR kit version
3.0 and a PCR Thermal Cycler Dice (Takara Biotechnology Co., Ltd.).
The primer sequences were as follows: RhoC upstream,
5′-ATGGCTGCAATCCGAAAG AAG-3′ and downstream,
5′-TCAGAGAATGGGACAGCCCCT-3′. Tubulin upstream,
5′-CACCCGTCTTCAGGGCTTCTTGGTTT-3′ and downstream,
5′-CATTTCACCATCTGGTTGGCTGGCTC-3′ (Sangon Biotech Co., Ltd.,
Shanghai, China). The PCR reaction mixture (50 µl) consisted
of: 1 µl cDNA, 10 µl 5X PCR buffer, 1 µl
upstream primer, 1 µl downstream primer, 4 µl
MgCl2, 1 µl mixed dNTP, 31.5 µl
ddH2O, and 0.5 µl Ex Taq DNA polymerase.
The annealing temperature was 57°C, and 30 cycles were set for RhoC
amplification and 25 cycles for tubulin amplification. Tubulin was
used as a housekeeping gene. Agarose gel electrophoresis was
conducted, then grayscale analysis was performed using the Image
Master Analysis system. The following equation was used to
calculate the inhibition ratio: Inhibition ratio = (grayscale in
the control group − grayscale in the experimental group) /
grayscale in the control group × 100%.
Western blot analysis for protein
determination
The cells were lysed with protein lysis buffer
(Watson Biotechnology Co., Ltd., Beijing, China) and RhoC protein
was determined through western blot analysis. The total cell
protein sample obtained from the lysed cells was loaded onto wells
(30 µg protein for each lane) for SDS-PAGE. After 12%
SDS-PAGE at 120 V, transmembrane electrophoresis onto a
polyvinylidene difluoride membrane (GE Osmonics, Inc., Minnetonka,
MN, USA) was performed for 1.5–2 h under constant electrical flow,
with the electrical current (rnA)=gel area ×2. The membrane was
blocked with 5% skimmed milk powder overnight. Subsequently, the
membrane was probed with primary antibodies at 37°C for 2 h. The
membrane was then washed three times (10 min/wash) with
tris-buffered saline containing 0.05% (v/v) Tween 20 (TBST;
Sigma-Aldrich, St. Louis, MO, USA), followed by 1.5 h incubation at
37°C with secondary antibodies. The membrane was subsequently
washed a further three times with TBST and the antibodies were
visualized using an enhanced chemiluminescence (ECL) kit (LumiPico
ECL kit; Shanghai ShineGene Molecular Biotech, Inc., Shanghai,
China), and the membrane was exposed to X-ray irradiation according
to the manufacturer's instructions of the ECL kit. The exposed
X-ray film was scanned using Tanon Image Note (Tanon Science &
Technology Co., Ltd., Shanghai, China) for grayscale analysis, and
β-actin was adopted as an internal reference.
MTT assay of cell growth
The present study included five experimental groups,
which were termed the RNAi group, the RAPA group (culture medium
containing 9.14 mg/l RAPA) (18),
the RNAi + RAPA group (pU6mRFP RhoC-siRNA transfected 24 h after
RAPA administration), the Scramble group (transfected with pU6m
RFPscramble-siRNA) and the HL7702 normal hepatocyte cell (Shanghai
Institute of Biochemistry and Cell Biology Cell Bank) group. A
total of 2×103 cells/well were seeded into a 96-well
plate, with a total reaction system at 200 µl per well and
incubated for seven days. MTT (5 mg/ml; 10 µl;
Sigma-Aldrich) was added to each well, then the culture was
continued for 6 h prior to the detection. The culture medium was
discarded, dimethyl sulfoxide (Sigma-Aldrich) was added (100
µl/well) and then the solution was mixed with vortexing for
5 min. The absor-bance was measured by a microplate reader (ELx800;
Bio-Tek Instruments, Inc., Winooski, VT, USA) at 490 nm wavelength.
The experiment was performed in triplicate for each group.
Argyrophilic protein analysis of cell
proliferation
Cells from each group were seeded in 24-well culture
plates containing cover slips. The culture plates were placed in an
incubator at 37°C, with 5% CO2. When cultivated cells
grew to a mono-layer, the cover slips were removed and cells were
fixed in 95% ethanol. The samples were immersed in deionized water
for hydration; then, ~5 drops (20 µl/drop) of silver nitrate
solution (0.2 g gelatin, 10 ml 1% formic acid, 20 ml 50% silver
nitrate; Sigma-Aldrich) were added onto the cover slips under the
exclusion of light, followed by incubation at room temperature for
1 h. The cells were repeatedly washed with deionized water,
dehydrated with 95–100% alcohol, cleared with xylene and mounted
with neutral gum (Sigma-Aldrich). Single black particles in the
nucleus were counted under a light microscope (CKX41; Olympus,
Tokyo, Japan) and three horizons were randomly counted.
Wright's stain to detect cell
apoptosis
Cells in the logarithmic growth phase were
harvested, with ~1×106 cells in each group, and then
washed with PBS. Cell were adjusted to 1×105 cells/ml,
inoculated in a 12-well culture plate (1 ml/well), with a cover
slip at the bottom of each well. A total of 24 h subsequent to
incubation, each cover slip was washed carefully with 3 ml PBS
three times. The cell slides were then fixed with methanol for 3–5
min, stained with Wright's solution (Hartman-Leddon Company, Inc.,
Philadelphia, PA, USA) for 2 min, immersed into Wright's phosphate
buffer (Wright's solution:PBS, 1:2) dilution for 4–10 min and then
rinsed with distilled water. The cell slides were mounted with
neutral gum after open-air drying. Images were captured under a
microscope.
Soft agar colony formation assays
Cells in the logarithmic growth phase were adjusted
to a density of 1×103/ml. Agar (5%; Sigma-Aldrich) was
completely dissolved in a boiling water bath. A portion of the 5%
agar was transferred into a small beaker, cooled to 50°C and
rapidly mixed with nine portions of fresh pre-warmed culture medium
at 37°C. The mixture was immediately poured into 24-well culture
plates, with each well containing 0.8 ml 0.5% agar medium and then
solidified at room temperature. A 0.3% agar medium was prepared
from 9.4 ml cell suspension in a small beaker (incubated at 37°C)
by rapid mixing with 0.6 ml 5% agar at 50°C. The agar culture was
immediately poured into 24-well plates, which were covered with
agar at their base, with 0.8 ml medium in each well, and solidified
at room temperature. A total of 500 cells were added per well. The
plates were placed into an incubator at 37°C with 5% CO2
and saturated humidity for two weeks. The clone forming efficiency
was counted when cell clones were >75 µm in diameter or
>50 cells in a single clone were observed under an inverted
microscope. The following equation was used to calculate the clone
formation percentage: Clone formation percentage = number of clones
− forming/seeded cells ×100%.
Transwell assay for detection of cell
migration
Cells in the logarithmic growth phase were digested
with trypsin (Amresco LLC, Solon OH, USA) to prepare a cell
suspension of 4×106 cells/ml. A total of 200 µl
cell suspension was added into the upper chambers of a Transwell
plate (Shanghai Yu Bo Biological Technology Co., Ltd., Shanghai,
China), and 500 µl culture medium containing penicillin and
streptomycin was added into the lower chamber, followed by
incubation at 37°C for 24 h. The culture medium was discarded, and
non-migrating cells were collected with a cotton swab. Cells were
washed twice with PBS, fixed with 4% paraformaldehyde for 20 min
then flushed with distilled water. A total of 500 µl 0.1%
crystal violet was added into the chamber, incubated at 37°C for 30
min and then washed with distilled water. The microscope was used
to observe migrated cells. Three visual fields were randomly
selected for counting and capturing of images.
Statistical analysis
Values are expressed as the mean ± standard
deviation. Experiments were repeated three times. SPSS 13.0
software (SPSS, Inc., Chicago, IL, USA) was used for statistical
analysis, and the t-test as well as an analysis of variance were
performed. P<0.05 was considered to indicate a statistically
significant difference.
Results
RhoC gene expression and RNAi gene
silencing effects
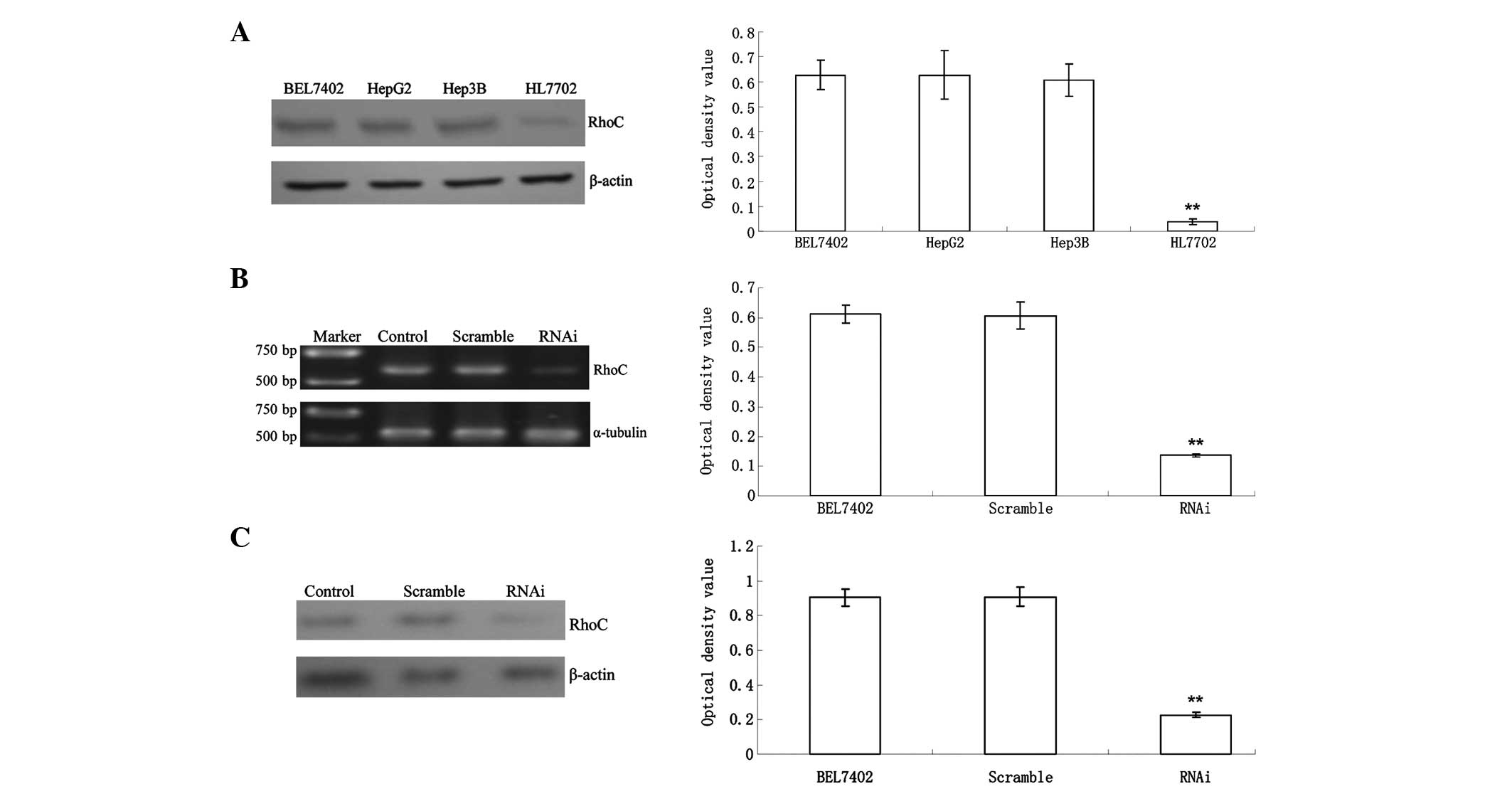
RhoC expression in the HL7702 normal human liver
cell line was significantly lower than that in BEL7402, HepG2 and
Hep3B hepatoma cells (0.04±0.01 vs. 0.63±0.06, 0.63±0.10 and
0.61±0.07; P<0.01) (Fig. 1A).
RhoC mRNA expression in the RNAi group decreased significantly
compared with that in the control and Scramble groups (0.26±0.02
vs. 0.90±0.05 and 0.91±0.06; P<0.01). The inhibition efficiency
was ~75% (Fig. 1B), suggesting
that RhoC-siRNA had a good gene silencing effect. Western blot
analysis revealed that RhoC protein expression in the RNAi group
was significantly lower than that in the control and scramble
group, with an inhibition ratio of 78.2% (P<0.01, Fig. 1C), in accordance with the PCR
results.
Combined RhoC knockdown and RAPA
treatment decrease the proliferation of hepatoma cells
From the 2nd day of RAPA treatment, the rate of cell
proliferation in the RNAi + RAPA group was significantly lower than
that in the RNAi, RAPA and Scramble groups (0.27±0.02 vs.
0.34±0.02, 0.37±0.02, 0.41±0.02; P<0.05), with no significant
difference compared with that in the normal HL7702 group (0.27±0.02
vs. 0.26±0.03; P>0.05) (Fig.
2A). Cell proliferation-associated gene expression revealed
that cyclin-dependent kinase 2 (CDK2) expression in the RNAi + RAPA
group was significantly lower than that in the RNAi, RAPA and
Scramble groups (0.10±0.02 vs. 0.52±0.03, 0.54±0.02 and 0.81±0.03;
P<0.01). No significant difference was observed between the RNAi
+ RAPA and HL7702 groups (0.10±0.02 vs. 0.07±0.01; P>0.05)
(Fig. 2B). P16 gene expression in
the RNAi + RAPA group was significantly higher than that in the
RNAi, RAPA and Scramble groups (0.72±0.04 vs. 0.27±0.04, 0.29±0.04
and 0.09±0.03; P<0.01), and no significant difference was found
when compared with the HL7702 group (0.72±0.04 vs. 0.77±0.05;
P>0.05) (Fig. 2B).
In the silver nitrate staining assay, the number of
stained particles in the nuclei in the RNAi + RAPA group was
significantly reduced when compared with that in the RNAi, RAPA and
Scramble groups (1.22±0.44 vs. 2.22±0.44, 2.44±0.53 and 3.78±0.67;
P<0.01) (Fig. 3). No notable
difference was found between the RNAi + RAPA and HL7702 groups
(1.22±0.44 vs. 1.33±0.50; P>0.05). Only a small number of
stained particles were observed in the RNAi + RAPA group,
indicating decreased cell proliferation, which was consistent with
the results of the MTT assay.
All of these results suggested that RhoC knockdown
and RAPA treatment had synergic effects in inhibiting hematoma cell
proliferation.
RhoC knockdown and RAPA treatment
increase the apoptotic rate and apoptosis-associated signaling in
hepatoma cells
Wright's staining revealed that the cells in the
Scramble group were homogeneous and that apoptosis was scarce. The
RNAi + RAPA, RNAi and RAPA groups contained a large number of
apoptotic cells, which displayed nuclear condensation,
fragmentation, deepened chromatin staining and wrinkled membranes,
with the apoptotic rate in the RNAi + RAPA group being higher than
that in the other experimental groups. The HL7702 cells also
exhibited a significant level of apoptosis. The examination of the
expression of apoptosis-associated genes demonstrated that B-cell
lymphoma-2 (Bcl-2) expression in the RNAi + RAPA group was
significantly lower than that in the RNAi, RAPA and Scramble groups
(0.09±0.02 vs. 0.32±0.05, 0.34±0.05 and 0.60±0.04; P<0.01), with
no significant difference compared with that in the HL7702 group
(0.09±0.02 vs. 0.08±0.03; P>0.05) (Fig. 4). Bcl-2-associated X protein (Bax)
gene expression in the RNAi + RAPA group was significantly higher
than that in the RNAi, RAPA and Scramble groups (0.64±0.08 vs.
0.31±0.04, 0.33±0.04 and 0.27±0.04; P<0.01). There was no
significant difference in Bax or Bcl-2 levels between the RNAi +
RAPA and HL7702 groups (0.64±0.08 vs. 0.53±0.07; P>0.05)
(Fig. 4).
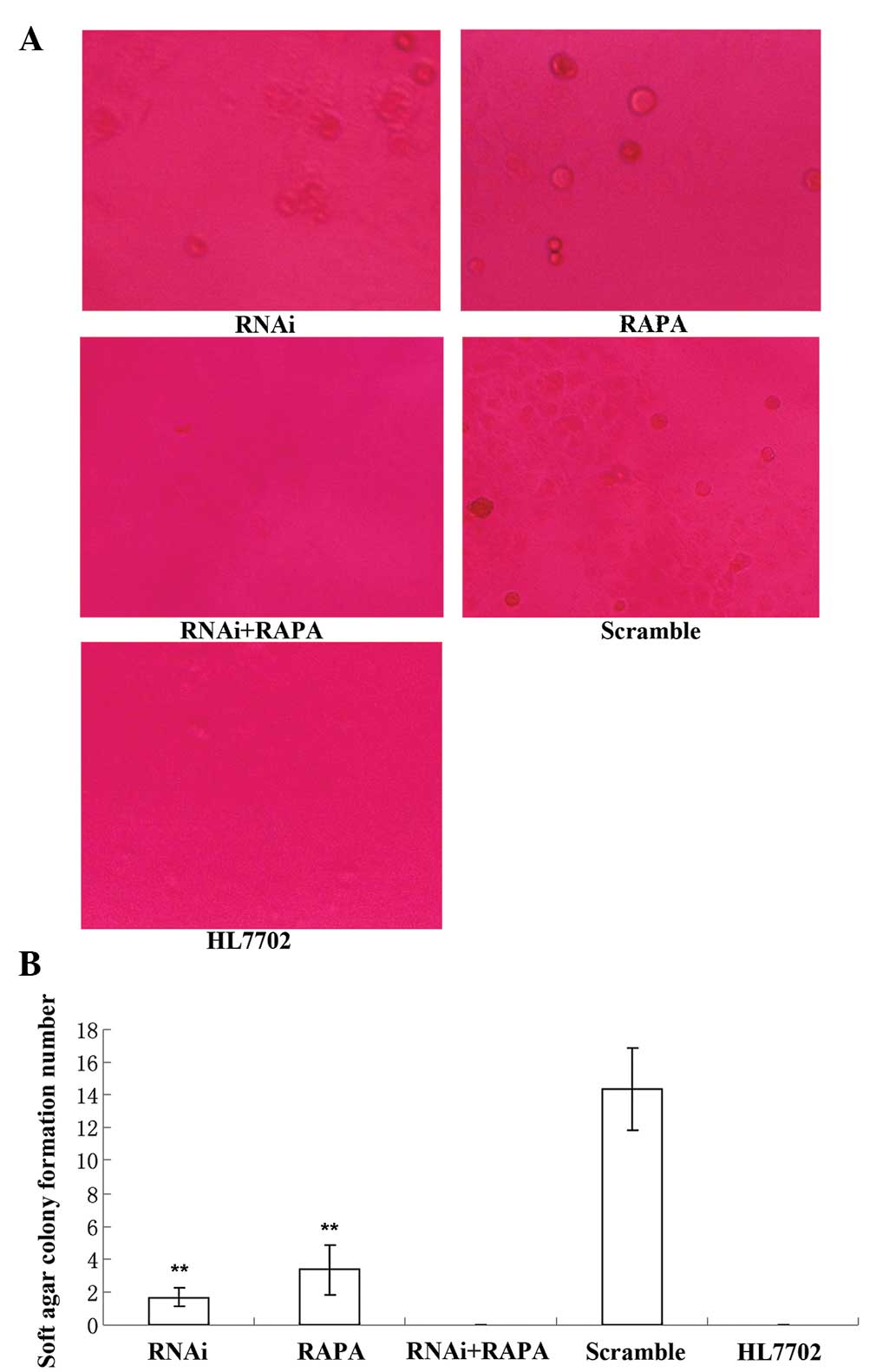
RhoC knockdown and RAPA treatment inhibit
cell colony formation of hepatoma cells
The soft agar assay showed that cell
colony-formation in the RNAi and RAPA groups was scarce, while no
colony formation was observed in the RNAi + RAPA and HL7702 groups
(Fig. 5). Soft agar cell colony
formation in the RNAi group was significantly lower than that in
the Scramble group (1.67±0.58 vs. 12.26±2.84; P<0.01), and the
difference between the RNAi and RAPA groups was not significant
(1.67±0.58 vs. 3.33±1.53; P>0.05) (Fig. 5).
RhoC knockdown and RAPA treatment
decrease cell migration as well as migration-associated protein
expression in hepatoma cells
The Transwell assay showed that cells which migrated
across the membrane in the RNAi, RAPA, RNAi + RAPA and HL7702
groups were rare and scattered, while in the Scramble group,
transmembrane cells were greater in number and dense in
distribution. Statistical analysis revealed that the number of
transmembrane cells in the RNAi + RAPA group was significantly
lower than that in the RNAi, RAPA and Scramble groups (35.33±3.51
vs. 65.33±6.51, 60.67±8.50 and 161.67±14.50; P<0.01), and no
significant difference was found when compared with the HL7702
group (35.33±3.51 vs. 25.67±5.86; P>0.05) (Fig. 6A). Detection of cell
migration-associated genes revealed that matrix metal-loproteinase
(MMP)-2 expression in the RNAi + RAPA group was significantly lower
than that in the RNAi, RAPA and Scramble groups (0.04±0.02 vs.
0.21±0.03, 0.20±0.04 and 0.67±0.04; P<0.01), and the difference
was not significant compared with that in the HL7702 group
(0.04±0.02 vs. 0.03±0.01; P>0.05). MMP-9 expression in the RNAi
+ RAPA group was markedly decreased compared with that in the RNAi,
RAPA and Scramble groups (0.06±0.02 vs. 0.23±0.02, 0.19±0.04 and
0.62±0.03; P<.0.01), and no significant difference was observed
when compared with that in the HL7702 group (0.06±0.02 vs.
0.03±0.01; P>0.05). VEGF expression in the RNAi + RAPA group was
also significantly lower than that in the RNAi, RAPA and Scramble
groups (0.07±0.02 vs. 0.24±0.04, 0.20±0.06 and 0.62±0.06;
P<0.01); while there was no significant difference compared with
that in the HL7702 group (0.07±0.02 vs. 0.04±0.02; P>0.05)
(Fig. 6B). These results
demonstrated that RhoC gene silencing combined with RAPA treatment
exerted marked migration-inhibitory effects on hepatocellular
carcinoma cells.
Discussion
Liver cancer results from the imbalance of cell
proliferation and apoptosis, involving complex biological processes
in which cell signaling pathways are important (19). To examine the combined effects of
inhibiting the RhoC/RhoC kinase (ROCK) and PI3K/Akt/mTOR pathways
on hepatocellular carcinoma cell growth, RhoC gene silencing and
RAPA treatment were applied. It was identified that inhibition of
the two pathways had synergistic effects and more effectively
inhibited the proliferation, metastasis and apoptosis of hepatoma
cells as compared with monotreatment.
RhoC, as a Rho subfamily member, is an important
intracellular signal transduction molecule. Extracellular stimulus
signals are able to promote the Rho guanosine diphosphate-bound
form of the protein from the inactive state into the active
guanosine triphosphate-bound state, affecting the cell biological
character by regulating the metabolism of tumor cell growth,
invasion and migration (20). RhoC
regulates the cytoskeleton through actin stress fibers and focal
adhesions, thereby affecting cell migration, invasion and other
functions. In a number of cancer types, including colorectal,
liver, breast and prostate cancer, abnormal expression of RhoC is
necessary, and is important in tumor growth, invasion and
metastasis (21). The present
study identified RhoC overexpression in three types of hepatoma
cell. By contrast, RhoC expression was low in normal liver cells,
consistent with the literature (19). A previous study by our group
reported that RhoC gene silencing was able to inhibit tumor cell
growth, migration and invasion, and promoted tumor cell apoptosis
(7).
The PI3K/AKT/mTOR signaling pathway has been widely
elucidated. It is involved in intracellular signal transduction of
several growth factors, and is closely associated with the
biological function of certain diseases (13). mTOR is the core molecule of this
pathway, and its sustained activation is associated with a variety
of tumor behaviors, including invasion and metastasis, tumor
recurrence, and patient prognosis (9). mTOR consists of two distinct
multiprotein complexes: mTORC1 and mTORC2. RAPA is sensitive to
mTORC1 and can markedly inhibit its activities. mTORC1 activates
S6K1 and 4EBP1, which are associated with mRNA translation. Diverse
stimuli can activate mTORC1, such as growth factors, nutrients,
energy and stress signals, and essential signaling pathways, such
as PI3K, mitogen-activated protein kinases and AMP-activated
protein kinase, in order to control cell growth, proliferation and
survival (22). The RhoC gene
silencing experiments in combination with RAPA administration
achieved greater effects than monotreatment. As RAPA is an mTOR
inhibitor, it is able to inhibit the PI3K/Akt/mTOR pathway, has a
marked cytostatic effect, and, in combination with RhoC gene
silencing, produced a synergistic effect, markedly inhibiting liver
cancer cell proliferation and migration, as well as promoting
apoptosis.
Normal operation of the cell cycle requires the
joint action of CDKs and cyclins. Certain CDKs and cyclins are also
molecular targets for cancer therapy. Studies have attempted to
interfere with these regulatory molecules for the treatment of
tumors (23). To examine the
mechanism of cell growth inhibition, the present study selected two
cell cycle-associated molecules to investigate. One was CDK2, which
promotes cell growth, and the other was P16, which inhibits cell
growth. The results revealed that RhoC gene silencing combined with
RAPA administration markedly decreased CDK2 expression and greatly
increased P16 expression. The observed changes in these two
important cell cycle regulatory molecules are known to hamper cell
cycle progression between the G1 and S phase, thereby inhibiting
cell proliferation. Silver nitrate staining confirmed the improved
inhibitory effect of the combined efforts. In addition, the
combined effects also changed the characteristics of liver cancer
growth, rendering hepatocellular carcinoma cells unable to grow in
soft agar.
Local spread and distant metastasis of cancer cells
is the major cause of mortality in patients with hepatocellular
carcinoma. In the process of tumor invasion and metastasis, the
extracellular basement membrane is an important target, other than
tumor angiogenesis. Tumor invasion of the basement membrane can be
divided into three steps. Firstly, cell adhesion to the basement
membrane; secondly, extracellular matrix (ECM) degradation, and
finally the migration of cells. The ECM is a complex structure,
composed of a variety of constituents, predominantly type IV
collagen, fibronectin and laminin, amongst others. It is a natural
barrier for tumor cell invasion and metastasis (24). Tumor cell invasion and metastasis
must penetrate the barrier of the ECM. The Transwell chamber
experiment is able to simulate the process of tumor cell invasion.
Aggressive cells adhere, deform and move through the bottom of the
filter chamber, demonstrating the ability of tumor cells to invade
and migrate through the basement membrane. RAPA combined with RhoC
gene silencing decreased the capacity of tumor cells to pass
through the Transwell membrane, indicating a reduced invasiveness
of the hepatoma cells.
Damage to the ECM barrier is the most important
process in tumor cell invasion and metastasis. Tumor cells are
activated by adhering to the matrix through cell surface receptors,
and secreting various proteolytic enzymes. Matrix
metalloproteinases (MMPs) are the main connective tissue-degrading
enzymes. ECM and the main structural protein of the basement
membrane are composed of type IV collagen, and MMP-2 and MMP-9 are
the most important enzymes for type IV collagen degradation,
involving the whole process of basement membrane degradation. A
previous study found that MMP-7, MMP-9 and tissue inhibitor of
metalloproteinase expression in non-small cell lung cancer were
significantly higher than that in the surrounding tissues (24). The activated PI3K/Akt/mTOR
signaling pathway may also upregulate MMP gene expression (25). It was hypothesized that RhoC gene
silencing combined with RAPA administration may decrease MMP
expression and activity, and suppress ECM degradation and tumor
cell migration, thus preventing tumor cell invasion and
metastasis.
VEGF is a vascular endothelial-specific mitogen,
regulating vascular endothelial cell proliferation and migration.
Although it stimulates tumor angiogenesis, it increases vascular
permeability and plasma protein extravasation, providing a matrix
for cancer metastasis. MMP-9 promotes the migration of vascular
endothelial cells, involving the process of tumor
neovascularization through the degradation of the basement membrane
and the ECM. Following specific binding with its receptor, VEGF is
able to facilitate vascular cell proliferation, increasing vascular
permeability and changing endothelial cell gene expression, and
thereby enhancing MMP synthesis (26). According to the present
experimental results, it is reasonable to hypothesize that RhoC
gene silencing and RAPA administration are able to suppress the
VEGF-induced tumor vasculature, thereby inhibiting liver cancer
cell invasion and metastasis, and their combination can further
enhance this inhibitory effect.
An important cause of the fatalities in patients
with liver cancer is chemical therapy resistance, which is closely
associated with a loss of balance of Bcl-2/Bax genes (27). RAPA administration combined with
RhoC gene silencing inhibited Bcl-2 expression and promoted Bax
expression. Bcl-2 is one of the most well known
apoptosis-associated genes, which is closely linked with cancer.
The Bcl-2 family consists of two groups of genes with antagonistic
functions. Bcl-2, Bcl-like protein 2 and Bcl extra large suppress
apoptosis, while Bax, Bcl-2-associated death promoter and BH3
interacting-domain death agonist promote apoptosis. Bcl-2 affects
tumor development by inhibiting cell apoptosis and prolonging cell
survival. The ratio of intracellular Bcl-2/Bax regulates cell
apoptosis. Bax homodimer formation can induce cell apoptosis. With
elevated Bcl-2 expression, Bax homodimer separates, combining with
Bcl-2 to form a Bax-Bcl-2 heterodimer, which is more stable than
the Bax-Bax homodimer, thereby offsetting the Bax-induced
apoptosis. Wright's staining morphologically confirmed that RAPA
administration combined with RhoC silencing had enhanced
pro-apoptotic effects on hepatoma cells.
In conclusion, the present study demonstrated that
silencing of RhoC combined with RAPA administration was able to
inhibit the rapid proliferation of hepatocellular carcinoma cells,
and deter tumor cell invasion and metastasis. Its mechanism may be
associated with the synergistic effects of inhibiting the Rho/ROCK
and PI3K/Akt/mTOR signaling pathways in liver cancer cells.
References
|
1
|
Abdel-Rahman O: Systemic therapy for
hepatocellular carcinoma (HCC): From bench to bedside. J Egypt Natl
Canc Inst. 25:165–171. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Harrold JM, Ramanathan M and Mager DE:
Network-based approaches in drug discovery and early development.
Clin Pharmacol Ther. 94:651–658. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhao Y, Zheng HC, Chen S, Gou WF, Xiao LJ
and Niu ZF: The role of RhoC in ovarian epithelial carcinoma: A
marker for carcinogenesis, progression, prognosis and target
therapy. Gynecol Oncol. 130:570–578. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Islam M, Sharma S, Kumar B and Teknos TN:
Atorvastatin inhibits RhoC function and limits head and neck cancer
metastasis. Oral Oncol. 49:778–786. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wu Y, Chen YC, Sang JR and Xu WR: RhoC
protein stimulates migration of gastric cancer cells through
interaction with scaffold protein IQGAP1. Mol Med Rep. 4:697–703.
2011.PubMed/NCBI
|
|
6
|
Wang W, Yang LY, Huang GW, Lu WQ, Yang ZL,
Yang JQ and Liu HL: Genomic analysis reveals RhoC as a potential
marker in hepatocellular carcinoma with poor prognosis. Br J
Cancer. 90:2349–2355. 2004.PubMed/NCBI
|
|
7
|
Xie S, Zhu M, Lv G, Zhang Q and Wang G:
The role of RhoC in the proliferation and apoptosis of
hepatocellular carcinoma cells. Med Oncol. 29:1802–1809. 2012.
View Article : Google Scholar
|
|
8
|
Xie S, Zhu M, Lv G, Geng Y, Chen G, Ma J
and Wang G: Overexpression of Ras homologous C (RhoC) induces
malignant transformation of hepatocytes in vitro and in nude mouse
xenografts. PLoS One. 8:e544932013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Psyrri A, Arkadopoulos N, Vassilakopoulou
M, Smyrniotis V and Dimitriadis G: Pathways and targets in
hepatocellular carcinoma. Expert Rev Anticancer Ther. 12:1347–1357.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yang H, Rudge DG, Koos JD, Vaidialingam B,
Yang HJ and Pavletich NP: mTOR kinase structure, mechanism and
regulation. Nature. 497:217–223. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Diaz-Padilla I, Duran I, Clarke BA and Oza
AM: Biologic rationale and clinical activity of mTOR inhibitors in
gynecological cancer. Cancer Treat Rev. 38:767–775. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sendur MA, Aksoy S, Zengin N and Altundag
K: Comparative efficacy study of 5-year letrozole or anastrozole in
postmenopausal hormone receptor-positive early breast cancer. J
BUON. 18:838–844. 2013.PubMed/NCBI
|
|
13
|
Khan KH, Yap TA, Yan L and Cunningham D:
Targeting the PI3K-AKT-mTOR signaling network in cancer. Chin J
Cancer. 32:253–265. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yamanaka K, Petrulionis M, Lin S, Gao C,
Galli U, Richter S, Winkler S, Houben P, Schultze D, Hatano E, et
al: Therapeutic potential and adverse events of everolimus for
treatment of hepatocellular carcinoma-systematic review and
meta-analysis. Cancer Med. 2:862–871. 2013. View Article : Google Scholar
|
|
15
|
Finn RS: Current and future treatment
strategies for patients with advanced hepatocellular carcinoma:
Role of mtor inhibition. Liver Cancer. 1:247–256. 2012. View Article : Google Scholar
|
|
16
|
Ruth MC, Xu Y, Maxwell IH, Ahn NG, Norris
DA and Shellman YG: RhoC promotes human melanoma invasion in a
PI3K/Akt-dependent pathway. J Invest Dermatol. 126:862–868. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lehman HL, Van Laere SJ, van Golen CM,
Vermeulen PB, Dirix LY and van Golen KL: Regulation of inflammatory
breast cancer cell invasion through Akt1/PKBα phosphorylation of
RhoC GTPase. Mol Cancer Res. 10:1306–1318. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Jin Z: Experimental research on the
combined effects of RhoC-siRNA and rapamycin in hepatocellular
carcinoma cell line. Master Thesis of Jilin University. 34–36.
2010.
|
|
19
|
Grise F, Bidaud A and Moreau V: Rho
GTPases in hepatocellular carcinoma. Biochim Biophys Acta.
1795:137–151. 2009.PubMed/NCBI
|
|
20
|
Wilson KF, Erickson JW, Antonyak MA and
Cerione RA: Rho GTPases and their roles in cancer metabolism.
Trends Mol Med. 19:74–82. 2013. View Article : Google Scholar :
|
|
21
|
Karlsson R, Pedersen ED, Wang Z and
Brakebusch C: Rho GTPase function in tumorigenesis. Biochim Biophys
Acta. 1796:91–98. 2009.PubMed/NCBI
|
|
22
|
Populo H, Lopes JM and Soares P: The mTOR
signalling pathway in human cancer. Int J Mol Sci. 13:1886–1918.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Diaz-Moralli S, Tarrado-Castellarnau M,
Miranda A and Cascante M: Targeting cell cycle regulation in cancer
therapy. Pharmacol Ther. 138:255–271. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Safranek J, Pesta M, Holubec L, Kulda V,
Dreslerova J, Vrzalova J, Topolcan O, Pesek M, Finek J and Treska
V: Expression of MMP-7, MMP-9, TIMP-1 and TIMP-2 mRNA in lung
tissue of patients with non-small cell lung cancer (NSCLC) and
benign pulmonary disease. Anticancer Res. 29:2513–2517.
2009.PubMed/NCBI
|
|
25
|
Wu MH, Lo JF, Kuo CH, Lin JA, Lin YM, Chen
LM, Tsai FJ, Tsai CH, Huang CY and Tang CH: Endothelin-1 promotes
MMP-13 production and migration in human chondrosarcoma cells
through FAK/PI3K/Akt/mTOR pathways. J Cell Physiol. 227:3016–3026.
2012. View Article : Google Scholar
|
|
26
|
Chaudhary AK, Pandya S, Ghosh K and
Nadkarni A: Matrix metalloproteinase and its drug targets therapy
in solid and hematological malignancies: An overview. Mutat Res.
753:7–23. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Liu Z, Cheng M and Cao M: Potential
targets for molecular imaging of apoptosis resistance in
hepatocellular carcinoma. Biomed Imaging Interv J.
7:e52011.PubMed/NCBI
|