Introduction
Colorectal cancer is one of the most common
malignancies and is a major health burden worldwide (1,2).
During the past decade, most studies have focused on the roles of
nuclear DNA alterations in colorectal cancer, while alterations in
mitochondrial DNA (mtDNA) have received a lesser amount of
attention. Human mtDNA is a 16,569-base circular double-stranded
DNA molecule, which contains genes coding for 13 polypeptides
involved in the respiratory chain, 22 transfer RNAs and 2 ribosomal
RNAs that are critical for protein synthesis in mitochondria
(3). Replication of mtDNA as well
as transcription and translation of mtDNA-coding genes are thought
to be the major alterations of mtDNA.
Replication of mtDNA induces alterations in the
mtDNA content, which is quantified by its copy number. The mtDNA
copy number varies from hundreds to >10,000 per cell (4). Previous studies have demonstrated
that increased mtDNA copy number was closely associated with an
increased mortality in advanced gastric cancer patients (5). Furthermore, increased mtDNA content
was shown to be associated with a decreased survival rate of
patients with tumors (5,6). A previous study by our group
demonstrated that the mtDNA copy number was increased in colorectal
cancer, and that the mtDNA copy number in clinicopathological
stages I and II was significantly higher than that in stages III
and IV, which indicated that the mtDNA copy number has a crucial
role during the initiation and progression of colorectal cancer
(7). Despite the involvement of
changes in the mtDNA content in the tumorigenesis of numerous
malignancies, the factors involved in the alterations of the mtDNA
content in colorectal cancer have largely remained elusive.
Alterations in gene transcription and translation
may lead to functional deficiency of mitochondria by de-regulating
their energy metabolism, leading to excessive generation of free
radicals, which trigger programmed cell death (8). Nicotinamide adenine dinucleotide
(NADH) is a rate-limiting enzyme of oxidative phosphorylation, and
nicotinamide adenine dinucleotide subunit 2 (ND-2) is encoded by
mtDNA. Although a previous study by our group has revealed that the
expression of ND-2 was increased in colorectal cancer (9), further research is required to
determine the accurate mechanism.
mtDNA contains a unique 1,124-bp non-coding region,
which is known as the displacement loop (D-loop). It has been
observed that DNA methylation occurs in the D-loop region of
mammals; however, D-loop regions of certain tumor tissue types were
found to be de-methylated (9-11).
Numerous studies have demonstrated that DNA hypomethylation is
typically associated with gene activation. Furthermore, DNA
hypomethylation also enhances DNA replication (12). However, whether DNA hypomethylation
of the D-loop region of mtDNA is involved in the regulation of the
mtDNA copy number and ND-2 expression in colorectal cancer has
remained elusive. In the present study, the correlation between the
methylation status of the D-loop region, mtDNA copy number and ND-2
expression was investigated in 65 colorectal cancers and their
corresponding non-cancerous tissues. In addition, a de-methylation
experiment was performed on the Caco-2 colorectal cancer cell line
by using 5-aza-2′-deoxycytidine (5-Aza).
Materials and methods
Patients and specimens
Colorectal cancer tissues and their corresponding
non-cancerous tissues were surgically resected from 30 colon- and
35 rectal cancer patients between November 2012 and January 2014 at
West China Hospital, Sichuan University (Chengdu, China). This
study was approved by the ethics committee of West China Hospital,
Sichuan University (Chengdu, China). Informed consent was obtained
from all patients. 36 of the patients were male and 25 were female
(age, 35–70 years; mean age, 53.8±12.3 years). All specimens were
immediately frozen in liquid nitrogen and stored at −80°C for
further DNA and protein analysis. Colorectal cancer tissues were
resected from the edge of the tumors. In addition, their
corresponding non-cancerous tissues at least 5 cm away from the
tumors were also collected. The tumor, nodes and metastasis
classification of malignant tumors (13) was applied to determine the
clinicopathological stages of colorectal cancer by a senior
pathologist.
Caco-2 cell culture and de-methylation
experiment
The Caco-2 human colon adenocarcinoma cell line
(Type Culture Collection of the Chinese Academy of Sciences,
Shanghai, China) was cultured and maintained in Dulbecco's modified
Eagle's medium (HyClone, Logan, UT, USA) in the presence of 10%
fetal bovine serum (FBS; HyClone) supplemented with 100 U
penicillin and streptomycin (HyClone). Cells were cultured at 37°C
in a humidified atmosphere containing 5% CO2. Caco-2
cells were serum-starved with 0.5% FBS and then incubated with 5 mM
5-Aza (Sigma-Aldrich, St. Louis, MO, USA) for 24 h.
Methylation-specific polymerase chain
reaction (MSP)
mtDNA was extracted from specimens and cells by
using the Mito isolation kit (GenMed Scientifics, Shanghai, China)
according to the manufacturer's instructions. Methylation of the
D-loop region of mtDNA was determined by MSP using
bisulfite-modified DNA, prepared using an EpiTect Bisulfite kit
(Qiagen, Dusseldorf, German), as previously described (9). Two primer sets were used to amplify
the D-loop region of mtDNA that incorporated a number of CpG sites,
one specific for the methylated sequence (D-loop-M forward,
5′-TGTTTCGGTTTTAGCGTTTC-3′ and reverse,
5′-TACTACTCTCCTCGCTCCGA-3′); and the other for the unmethylated
sequence (D-loop-U forward, 5′-GGGTGTTTTGGTTTTAGTGTTTT-3′ and
reverse, 5′-ATACTACTCTCCTCACTCCAAAC-3′). Primers were provided by
Invitrogen Life Technologies, Carslbad, CA, USA). Universal
methylated DNA (Qiagen) was used as a positive control and
double-distilled H2O was served as a negative control.
MSP assays were performed at least in duplicate.
Quantification of the mtDNA copy
number
The relative mtDNA copy number was measured by
real-time quantitative polymerase chain reaction (qPCR) as
previously described (7). Briefly,
two pairs of primers were employed, one primer pair was used for
amplification of the mtDNA (forward, 5′-TACTCACCAGACGCCTCAACCG-3′
and reverse, 5′-TTATCGGAATGGGAGGTGATTC-3′), and the other primer
pair for the β-actin gene was run in parallel to standardize the
input DNA (forward, 5′-CGGGAAATCGTGCGTGACAT-3′ and reverse,
5′-GAAGGAAGGCTGGAAGAGTG-3′). The qPCR amplification was performed
by using a SYBR Green qPCR kit (Bio-Rad Laboratories, Inc.,
Hercules, CA, USA) according to manufacturer's instructions on an
Eppendorf Mastercycler ep realplex (Eppendorf, Hamburg, Germany).
For the standard curve, an ultraviolet spectrophotometer (GE
Healthcare Life Sciences, Piscataway, NJ, USA) was applied to
quantify the initial concentration of the amplification products of
the mtDNA and β-actin. Subsequently, the amplification products
were serially diluted with a two-fold dilution to generate a
six-point standard. Each measurement was performed in triplicate
and a non-template control was included in each experiment.
Western blot analysis of ND-2
protein
Total protein was extracted from the tissues and
cells by using a protein extraction kit (Nanjing Kaiji, Nanjing,
China) following the manufacturer's instructions. After
quantification by using the bicinchoninic acid assay (Nanjing
Kaiji, Nanjing, China), 50 mg proteins were separated by SDS-PAGE
and transferred onto nitrocellulose membranes, which were incubated
with rabbit anti-human polyclonal antibodies against ND-2 (1:400;
cat. no. ab102753; Abcam, Cambridge, MA, USA) and cytochrome C
oxidase subunit IV (Cox IV; 1:400; cat. no. bs-10257R; Bioss,
Beijing, China) at 4°C overnight. The blots were washed and
incubated with appropriate horseradish peroxidase-conjugated
secondary antibodies (1:20,000; Santa Cruz Biotechnology, Inc.,
Santa Cruz, CA, USA) and visualized with an enhanced
chemiluminescence detection kit (Amersham Pharmacia, Uppsala,
Sweden). The autoradio-graphs were scanned following exposure of
the membranes to Kodak XAR film (Kodak, Rochester, NY, USA).
Protein expression was determined using Quantity One software 4.5.0
(Bio-Rad Laboratories, Inc.), and the protein expression levels
were normalized to Cox IV.
Statistical analysis
All data are expressed as the mean ± standard
deviation and were analyzed using SPSS 13.0 software (SPSS, Inc.,
Chicago, IL, USA). Student's t-test was applied to compare
differences in quantitative data. The χ2 test was
applied to analyze differences between methylation rates and
clinicopathological stages. P<0.05 was considered to indicate a
statistically significant difference between values.
Results
D-loop region is de-methylated in
colorectal cancer
The methylation status and methylation rate of the
D-loop region in the patient tissue samples, determined by the MSP
assay, are listed in Table I. The
results showed that the methylation rate of the D-loop region in
all 65 colorectal cancer tissues was markedly reduced compared with
that in their corresponding non-cancerous tissues (13.8 vs. 81.5%;
P<0.05). There was no significant correlation between the
methylation rate of the D-loop region and the age or gender in
colorectal cancer tissues and their corresponding non-cancerous
tissues. However, the methylation rate of the D-loop region in the
colorectal cancer tissues was markedly decreased in
clinicopathological stages III and IV compared with stages I and II
(7.1 and 0% vs. 25 and 16%; P<0.05). The methylation rate of the
D-loop region in non-cancerous tissues was similar among the four
clinicopathological stages (81.3, 79.2, 85.7 and 81.8%;
P>0.05).
 | Table IAssociation between methylation rate
of the D-loop region and clinicopathological parameters in 65
colorectal cancer cases. |
Table I
Association between methylation rate
of the D-loop region and clinicopathological parameters in 65
colorectal cancer cases.
| Characteristic | Number of cases | Methylated D loop, n
(%)
|
|---|
| Colorectal
cancer | Corresponding
non-cancerous |
|---|
| Total | 65 | 9 (13.8) | 53 (81.5) |
| Mean age (years) |
| <50 | 39 | 5 (12.8) | 32 (82.1) |
| ≥50 | 26 | 4 (15.3) | 21 (80.8) |
| Gender |
| Male | 36 | 6 (16.7) | 30 (83.3) |
| Female | 29 | 3 (10.3) | 23 (79.3) |
| Clinicopathological
stage |
| I | 16 | 4 (25) | 13 (81.3) |
| II | 24 | 4 (16.7) | 19 (79.2) |
| III | 14 | 1 (7.1) | 12 (85.7) |
| IV | 11 | 0 (0) | 9 (81.8) |
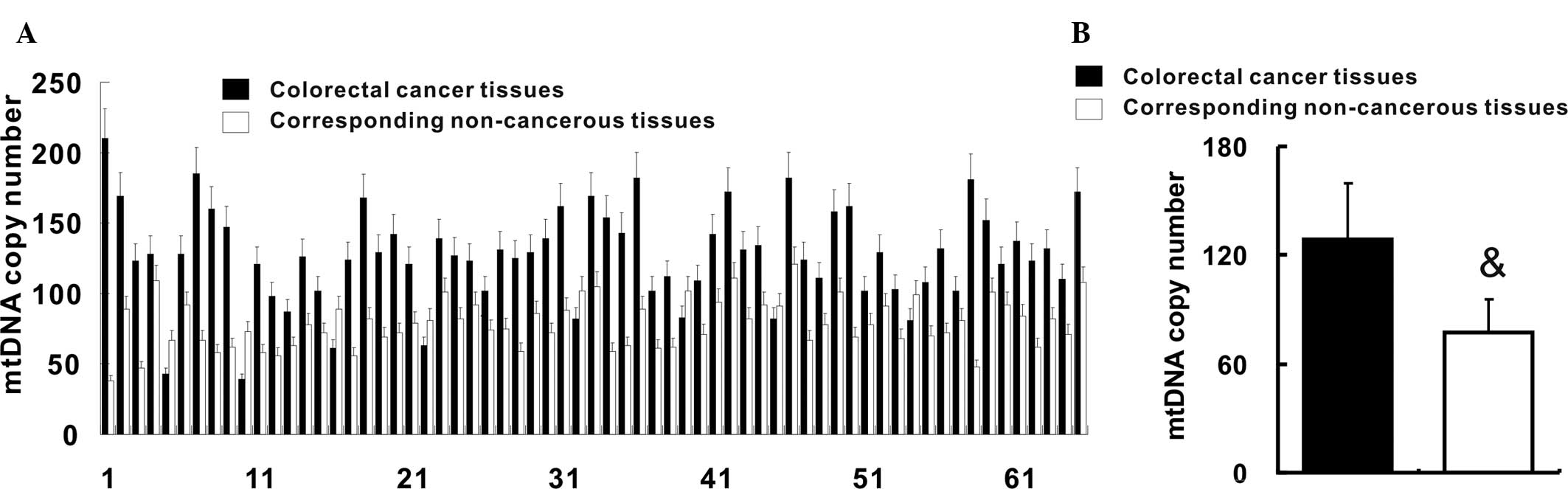
mtDNA copy number is increased in
colorectal cancer
qPCR was employed to quantify of the copy number of
mtDNA in colorectal cancer tissues and their corresponding
non-cancerous tissues. The relative mtDNA copy number was increased
in 61 (93.8%) colorectal cancer tissues when compared with that in
their corresponding non-cancerous tissues (Fig. 1A). In addition, the mean relative
mtDNA copy number in colorectal cancer tissues was increased when
compared with that in the corresponding non-cancerous tissues
(127.2±34.3 vs. 78.7±17.3; P<0.05) (Fig. 1B).
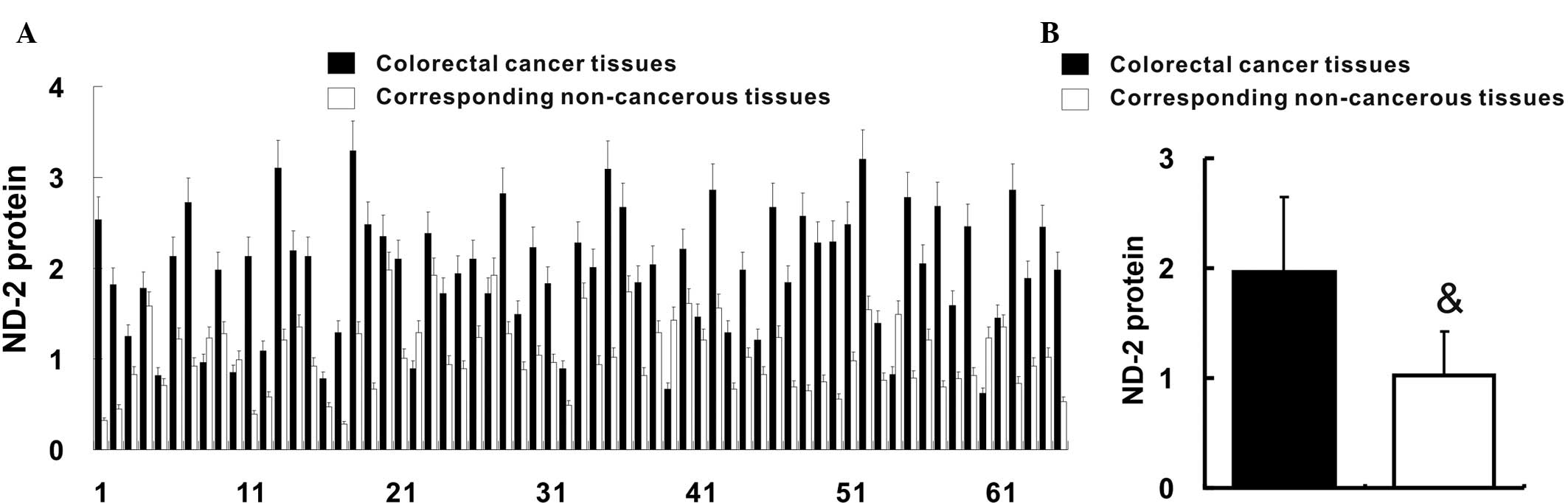
ND-2 expression is upregulated in
colorectal cancer
ND-2 protein in colorectal cancer tissues and their
corresponding non-cancerous tissues was quantified by western blot
analysis. The relative ND-2 protein content was increased in 58
(89.2%) colorectal cancer tissues when compared with that in their
corresponding non-cancerous tissues (Fig. 2A). In accordance with this result,
the average relative ND-2 protein content of colorectal cancer
tissues was also increased when compared with that in their
corresponding non-cancerous tissues (1.97±0.68 vs. 1.03±0.39;
P<0.05) (Fig. 2B).
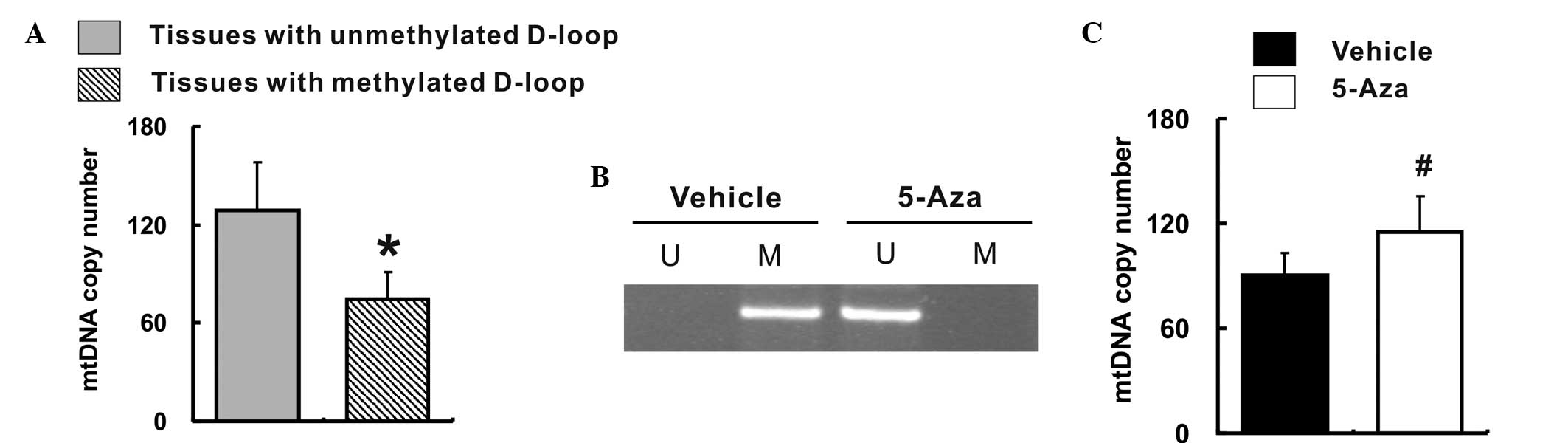
De-methylation of the D-loop region is
associated with an increased mtDNA copy number
In order to characterize the manner in which the
mtDNA copy number was altered, the correlation between the relative
mtDNA copy number and the methylation status of the D-loop region
was investigated in all 65 colorectal cancer tissues and their
corresponding non-cancerous tissues. Among the 130 tissue samples,
68 (52.3%) were unmethylated in the D-loop region, while 62 (47.7%)
tissues were methylated in the D-loop region. Furthermore, the
relative mtDNA copy number was markedly elevated in the tissues
with an unmethylated D-loop region when compared with the tissues
with a methylated D-loop region (128.8±29.7 vs. 76.4±14.6;
P<0.05), as shown in Fig. 3A.
Subsequently, the present study evaluated the effect of
de-methylation of the D-loop region in vitro. The MSP
results confirmed that treatment with 5-Aza altered the D-loop
region from a methylation to a de-methylation status (Fig. 3B). Consequently, the relative mtDNA
copy number was significantly increased in the 5-Aza-treated Caco-2
cells when compared with that measured in the vehicle-treated
Caco-2 cells (90.2±13.1 vs. 115.4±19.8; P<0.05), as shown in
Fig. 3C.
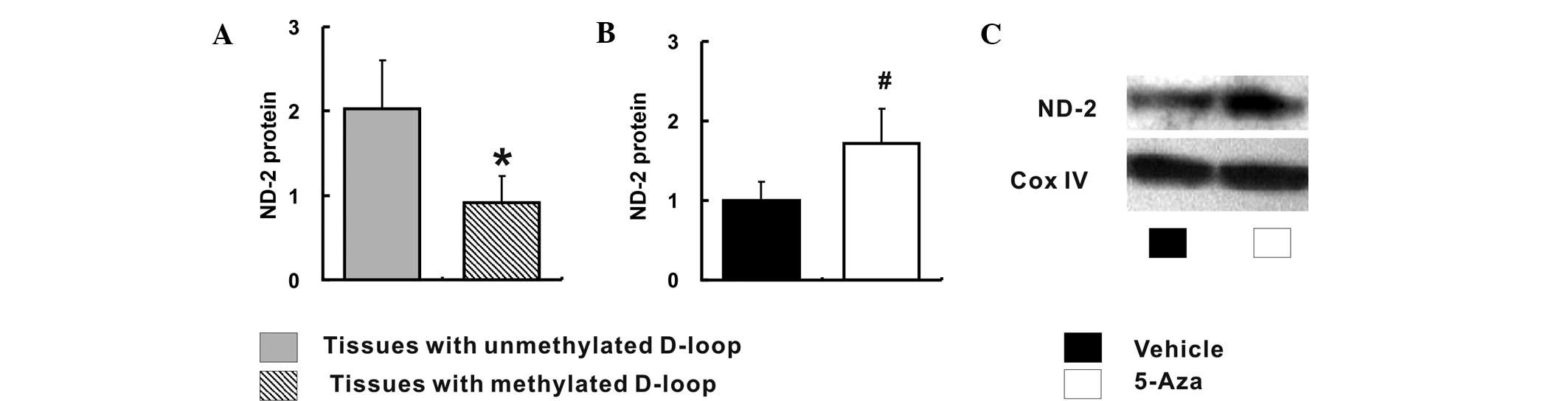
De-methylation of the D-loop region is
associated with increased ND-2 expression
The correlation between relative ND-2 expression and
methylation status of the D-loop region was also investigated. The
ND-2 expression was significantly increased in tissues with an
unmethylated D-loop region when compared to that in tissues with a
methylated D-loop region (2.03±0.58 vs. 0.92±0.31; P<0.05)
(Fig. 4A). Furthermore, the ND-2
expression was conspicuously increased in 5-Aza-treated Caco-2
cells when compared with that in the vehicle-treated Caco-2 cells
(1.00±0.23 vs. 1.72±0.43; P<0.05) (Fig. 4B and C).
Elevated mtDNA copy number is correlated
with increased ND-2 expression
The correlation between the mtDNA copy number and
the relative ND-2 expression in colorectal cancer tissues and their
corresponding non-cancerous tissues was investigated by using a
general linear correlation model. A positive linear correlation
between mtDNA copy number and relative ND-2 expression was observed
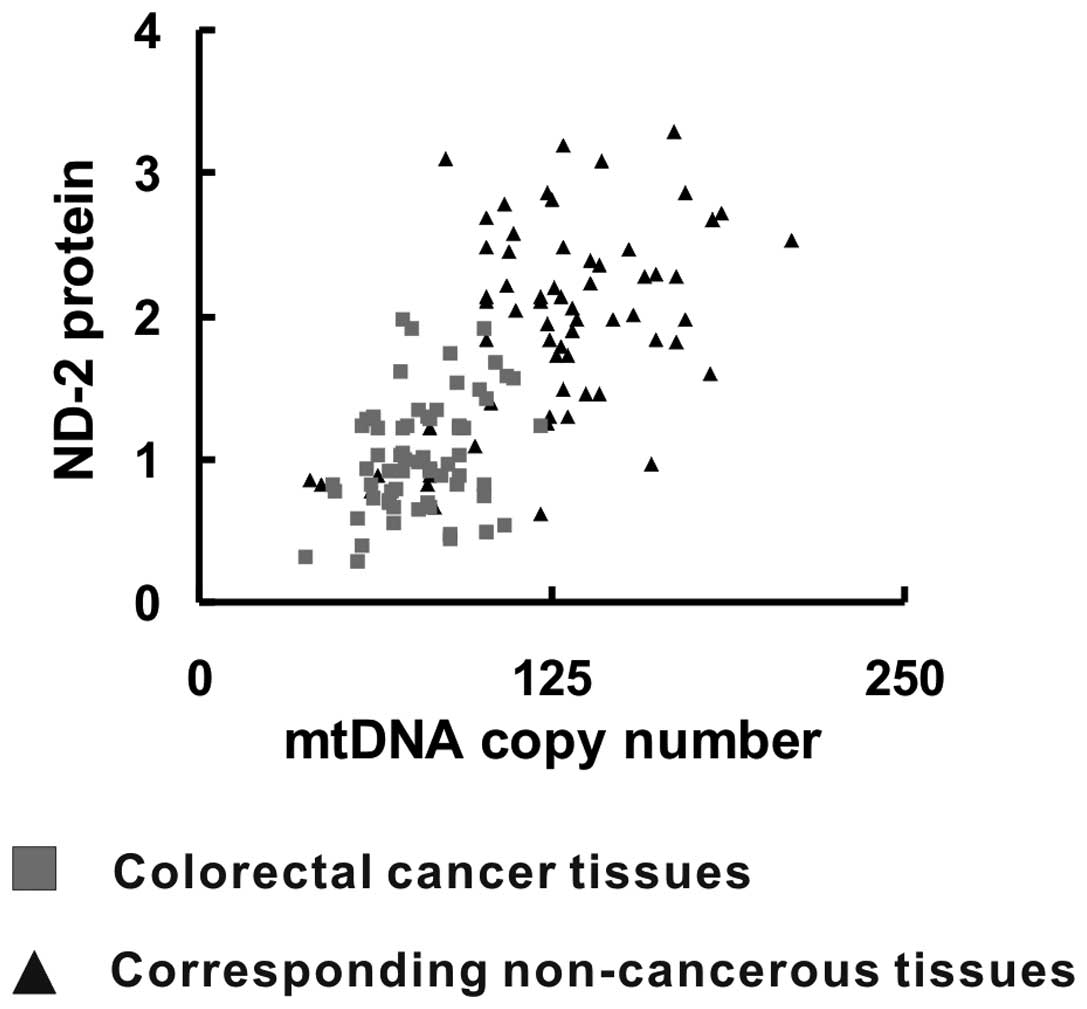
(R=0.685, P<0.05) (Fig. 5),
which indicated that increased ND-2 expression was correlated with
an elevated mtDNA copy number.
Discussion
Abnormal epigenetic alterations of several nuclear
DNA-encoded oncogenes have been suggested to be involved in driving
the tumorigenesis of colorectal cancer (14); however, little attention has been
paid to the involvement of mtDNA alternation in the pathobiology of
colorectal cancer. The present study demonstrated that
de-methylation of the D-loop region elevated the mtDNA copy number
and that the expression of ND-2 was concomitantly increased in
colorectal cancer tissues; furthermore, an association of the
methylation status of the D-loop region with the
clinicopathological stage was established in colorectal cancer
tissues. In addition, the de-methylation of the D-loop region was
associated with an elevated mtDNA copy number and an increased ND-2
expression in tissue samples as well as in the in vitro cell
experiment.
The epigenetic modification of the D-loop region
regulates the replication and the transcription of mtDNA (10,15).
Methylation, one of the most important types of epigenetic
modification, occurs at the CpG islands and is typically associated
with gene silencing (12). The
present study manifested that the de-methylation rate of the D-loop
region in colorectal cancer tissues was significantly higher than
that in the corresponding non-cancerous tissues. Furthermore, the
D-loop region was predominantly de-methylated in samples with
clinicopathological stages I and II (25 and 16.7%, respectively),
while it was largely or completely de-methylated in samples with
clinicopathological stages III and IV (7.1 and 0%). The gradient of
the de-methylation rate from clinicopathological stages I-IV
suggested that the de-methylation of the D-loop region increases
during the progression of colorectal cancer. These findings are
consistent with the results of a previous study, which indicated
that the de-methylation of the D-loop region is an early molecular
event in the genesis of colorectal cancer (9).
In post-mitotic cells, mtDNA replicates
continuously, leading to an increases in the mtDNA copy number
(16); however, the mechanisms of
mtDNA replication have remained to be fully elucidated. The D-loop,
a non-coding region of the mtDNA, is important for the regulation
of mitochondrial genome replication and expression, as it contains
the initial site of heavy-chain replication and the promoters for
heavy- and light-chain transcription (17). Mutation or epigenetic modifications
in the D-loop region may affect mtDNA replication and change the
expression pattern of mtDNA (10,15,18,19).
The present study manifested that de-methylation of the D-loop
region was associated with an elevated mtDNA copy number and
increased ND-2 expression in colorectal cancer tissues as well as
in their corresponding non-cancerous tissues. Of note, the mtDNA
copy number and ND-2 expression were significantly increased after
5-Aza treatment. These findings suggested that de-methylation of
the D-loop region is an important factor that regulates mtDNA
replication and ND-2 expression in colorectal cancer; however, the
exact mechanism remains elusive.
It has been gradually established that mtDNA
maintenance protein mitochondrial transcription factor A (TFAM)
regulates mtDNA levels (16,20).
TFAM stimulates transcription and replication of mtDNA by binding
to the D-loop region (21).
However, methylation of the CpG islands of the D-loop region
inhibits transcription factor binding, consequently suppressing
transcription and replication of mtDNA. In the present study,
methylation of the D-loop region was observed in 13.8% of
colorectal cancer tissues and 81.5% of their corresponding
non-cancerous tissues. Furthermore, de-methylation of the D-loop
region resulted in elevation of the mtDNA copy number and in
induction of ND-2 expression in vitro. From these results,
it can be presumed that de-methylation of the D-loop region
facilitates TFAM binding and eventually promotes the replication
and transcription of mtDNA. However, further study is required to
verify this hypothesis.
Cancer cells require higher levels of energy than
normal cells to sustain their rapid proliferation. The energy is
mainly supplied by oxidative phosphorylation in the mitochondria
(22,23). ND-2 is a subunit of NADH which is
involved in oxidative phosphorylation. In the present study,
upregulation of ND-2 was observed in colorectal cancer tissues
compared with that in their corresponding non-cancerous tissues.
Presumably, the increase of ND-2 gives rise to enhanced oxidative
phosphorylation in colorectal cancer. In accord with this result,
it was demonstrated by a recent study that circulating cancer cells
exhibit enhanced mitochondrial biogenesis and respiration, as well
as up-regulation of genes associated with oxidative phosphorylation
(22). Furthermore, the relative
ND-2 expression was linearly correlated with the mtDNA copy number,
which suggested that the elevated mtDNA copy number together with
the de-methylation of the D-loop region may be involved in the
up-regulation of ND-2 expression.
In conclusion, in colorectal cancer tissues, the
de-meth-ylation rate of the D-loop region, the mtDNA copy number
and ND-2 expression were significantly higher than those in the
corresponding non-cancerous tissues. De-methylation of the D-loop
region may be involved in the regulation of the mtDNA copy number
and ND-2 expression.
Acknowledgments
The present study was supported by a grant from the
Natural Science Fund of China (no. 81301770). The authors would
like to thank the doctors from the Gastrointestinal Surgery
Department of West China Hospital (Chengdu, China) for collecting
the specimens.
References
|
1
|
Siegel R, Desantis C and Jemal A:
Colorectal cancer statistics, 2014. CA Cancer J Clin. 64:104–117.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Center MM, Jemal A, Smith RA and Ward E:
Worldwide variations in colorectal cancer. CA Cancer J Clin.
59:366–378. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Grivell LA: Mitochondrial DNA. Small,
beautiful and essential. Nature. 341:569–571. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lightowlers RN, Chinnery PF, Turnbull DM
and Howell N: Mammalian mitochondrial genetics: Heredity,
heteroplasmy and disease. Trends Genet. 13:450–455. 1997.
View Article : Google Scholar
|
|
5
|
Zhang G, Qu Y, Dang S, Yang Q, Shi B and
Hou P: Variable copy number of mitochondrial DNA (mtDNA) predicts
worse prognosis in advanced gastric cancer patients. Diagn Pathol.
8:1732013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Cheau-Feng Lin F, Jeng YC, Huang TY, Chi
CS, Chou MC and Chin-Shaw Tsai S: Mitochondrial DNA copy number is
associated with diagnosis and prognosis of head and neck cancer.
Biomarkers. 19:269–274. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Feng S, Xiong L, Ji Z, Cheng W and Yang H:
Correlation between increased copy number of mitochondrial DNA and
clinicopatho-logical stage in colorectal cancer. Oncol Lett.
2:899–903. 2011.
|
|
8
|
Boland ML, Chourasia AH and Macleod KF:
Mitochondrial dysfunction in cancer. Front Oncol. 3:2922013.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Feng S, Xiong L, Ji Z, Cheng W and Yang H:
Correlation between increased ND2 expression and demethylated
displacement loop of mtDNA in colorectal cancer. Mol Med Rep.
6:125–130. 2012.PubMed/NCBI
|
|
10
|
Bellizzi D, D'Aquila P, Scafone T,
Giordano M, Riso V, Riccio A and Passarino G: The control region of
mitochondrial DNA shows an unusual CpG and non-CpG methylation
pattern. DNA Res. 20:537–547. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Roos-Araujo D, Stuart S, Lea RA, Haupt LM
and Griffiths LR: Epigenetics and migraine; complex mitochondrial
interactions contributing to disease susceptibility. Gene. 543:1–7.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Esteller M: CpG island hypermethylation
and tumor suppressor genes: A booming present, a brighter future.
Oncogene. 21:5427–5440. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Mates IN, Jinga V, Csiki IE, Mates D, Dinu
D, Constantin A and Jinga M: Single nucleotide polymorphisms in
colorectal cancer: associations with tumor site and TNM stage. J
Gastrointestin Liver Dis. 21:45–52. 2012.PubMed/NCBI
|
|
14
|
Matsubara N: Epigenetic regulation and
colorectal cancer. Dis Colon Rectum. 55:96–104. 2012. View Article : Google Scholar
|
|
15
|
Zhang R, Zhang F, Wang C, Wang S, Shiao YH
and Guo Z: Identification of sequence polymorphism in the D-Loop
region of mitochondrial DNA as a risk factor for hepatocellular
carcinoma with distinct etiology. J Exp Clin Cancer Res.
29:1302010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ylikallio E, Tyynismaa H, Tsutsui H, Ide T
and Suomalainen A: High mitochondrial DNA copy number has
detrimental effects in mice. Hum Mol Genet. 19:2695–2705. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Penta JS, Johnson FM, Wachsman JT and
Copeland WC: Mitochondrial DNA in human malignancy. Mutat Res.
488:119–133. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang J, Guo Z, Bai Y, Cui L, Zhang S and
Xu J: Identification of sequence polymorphisms in the displacement
loop region of mitochondrial DNA as a risk factor for renal cell
carcinoma. Biomed Rep. 1:563–566. 2013.
|
|
19
|
Tommasi S, Favia P, Weigl S, Bianco A,
Pilato B, Russo L, Paradiso A and Petruzzella V: Mitochondrial DNA
variants and risk of familial breast cancer: An exploratory study.
Int J Oncol. 44:1691–1698. 2014.PubMed/NCBI
|
|
20
|
Maniura-Weber K, Goffart S, Garstka HL,
Montoya J and Wiesner RJ: Transient overexpression of mitochondrial
transcription factor A (TFAM) is sufficient to stimulate
mitochondrial DNA transcription, but not sufficient to increase
mtDNA copy number in cultured cells. Nucleic Acids Res.
32:6015–6027. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kurita T, Izumi H, Kagami S, Kawagoe T,
Toki N, Matsuura Y, Hachisuga T and Kohno K: Mitochondrial
transcription factor A regulates BCL2L1 gene expression and is a
prognostic factor in serous ovarian cancer. Cancer Sci.
103:239–444. 2012. View Article : Google Scholar
|
|
22
|
LeBleu VS, O'Connell JT, Gonzalez Herrera
KN, Wikman H, Pantel K, Haigis MC, de Carvalho FM, Damascena A,
Domingos Chinen LT, Rocha RM, et al: PGC-1α mediates mitochondrial
biogenesis and oxidative phosphorylation in cancer cells to promote
metastasis. Nat Cell Biol. 16:992–1003. 2014. View Article : Google Scholar
|
|
23
|
Lim SH, Wu L, Kiew LV, Chung LY, Burgess K
and Lee HB: Rosamines targeting the cancer oxidative
phosphorylation pathway. PloS one. 9:e829342014. View Article : Google Scholar : PubMed/NCBI
|