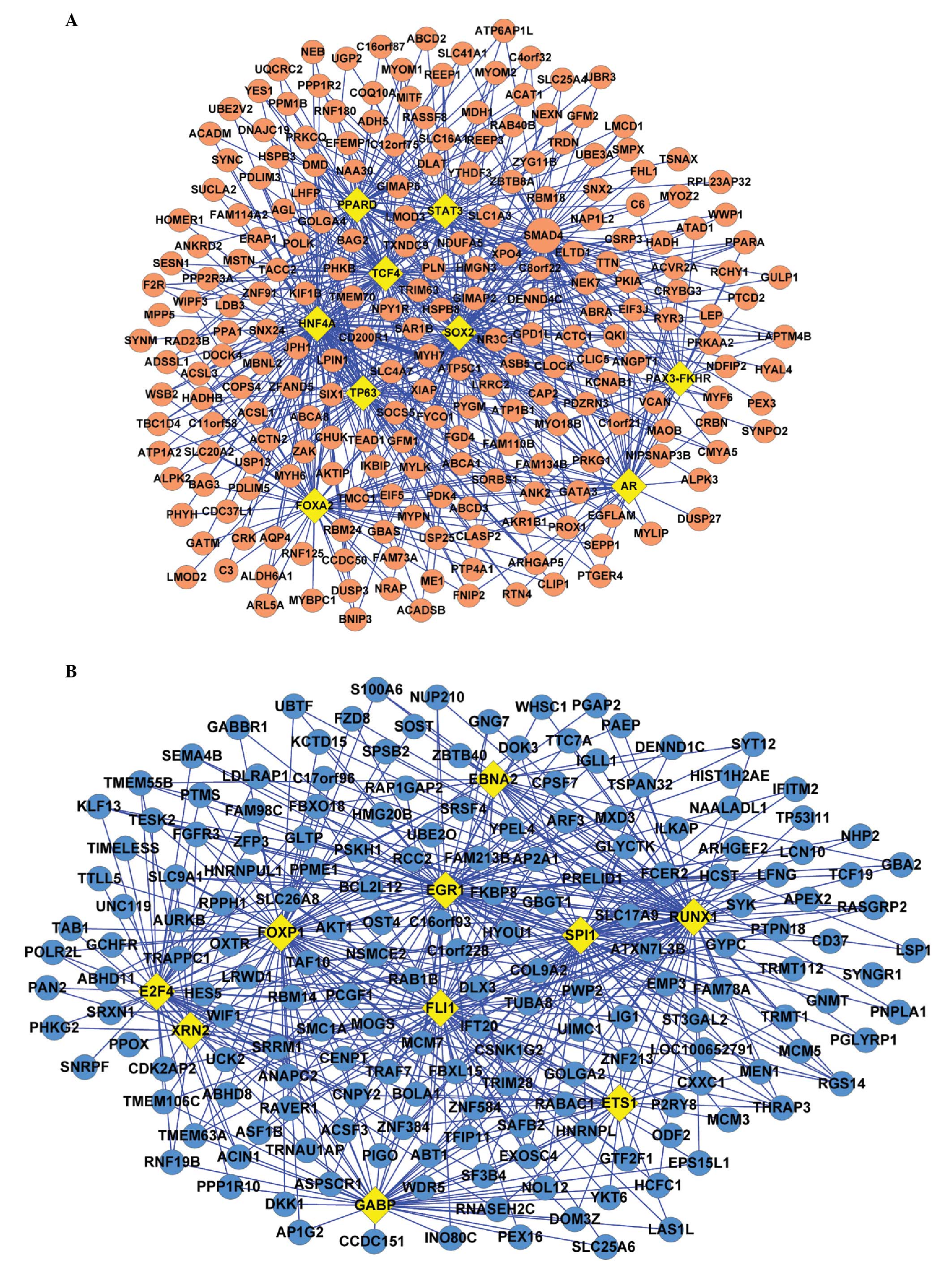
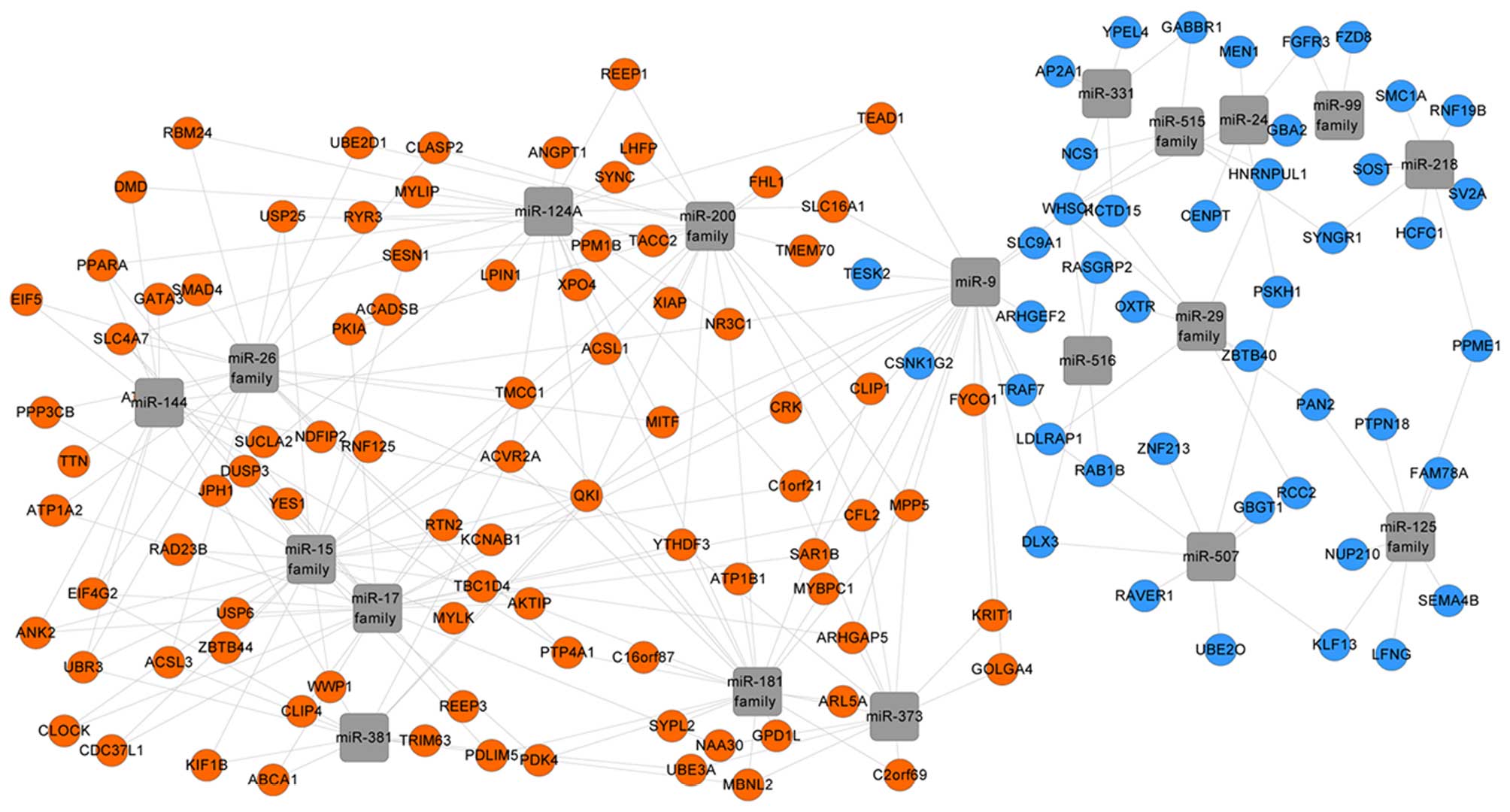
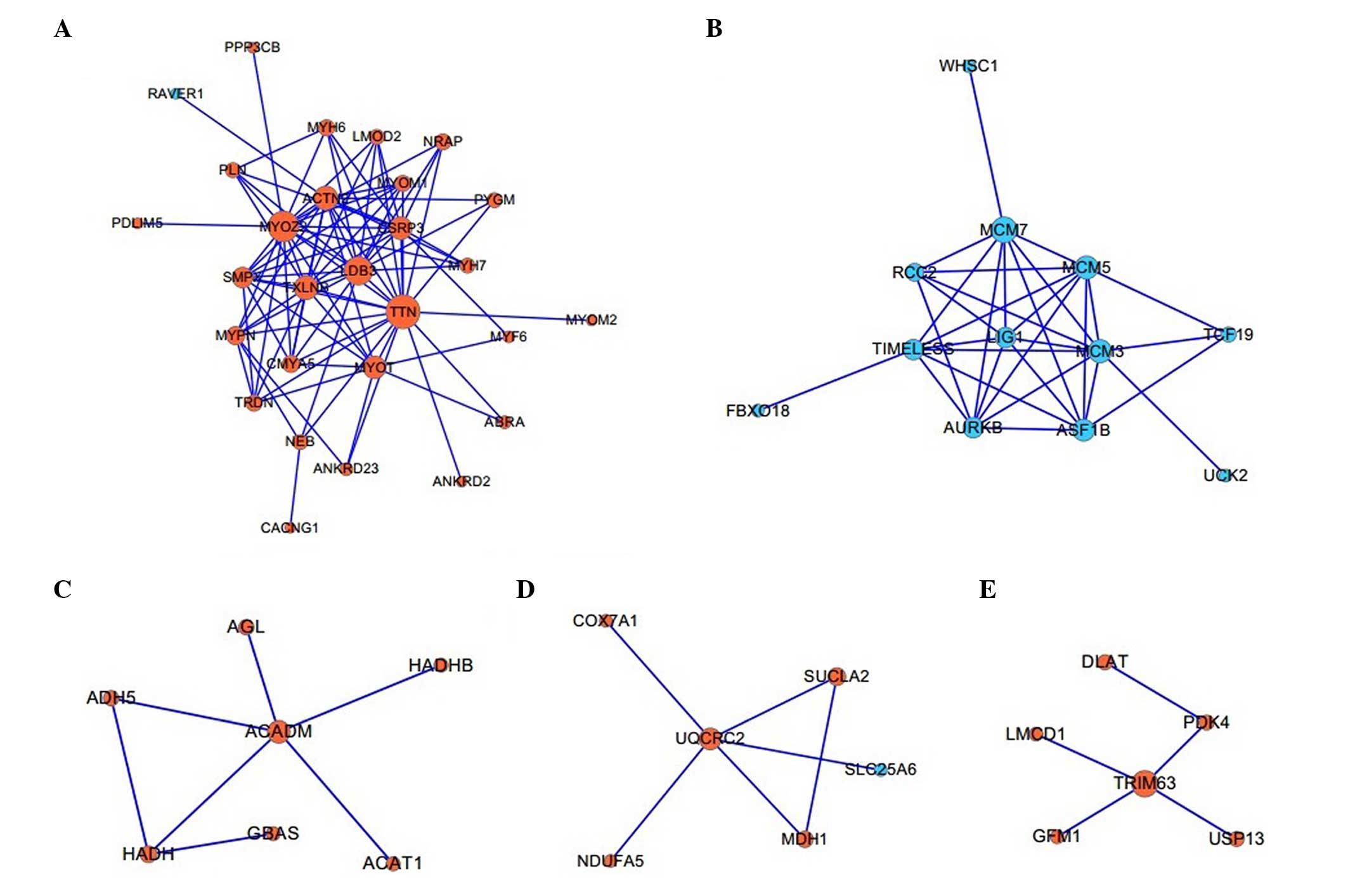
|
1
|
Bouillon R, Burckhardt P, Christiansen C,
et al: Consensus development conference: Prophylaxis and treatment
of osteoporosis. Am J Med. 90:107–110. 1991. View Article : Google Scholar
|
|
2
|
Marcus R: Post-menopausal osteoporosis.
Best Pract Res Clin Obstet Gynaecol. 16:309–327. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Michaëlsson K, Melhus H, Ferm H, Ahlbom A
and Pedersen NL: Genetic liability to fractures in the elderly.
Arch Intern Med. 165:1825–1830. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Mullin BH, Prince RL, Dick IM, Hart DJ,
Spector TD, Dudbridge F and Wilson SG: Identification of a role for
the ARHGEF3 gene in postmenopausal osteoporosis. Am J Hum Genet.
82:1262–1269. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Capulli M, Olstad OK, Önnerfjord P,
Tillgren V, Muraca M, Gautvik KM, Heinegård D, Rucci N and Teti A:
The C-terminal domain of chondroadherin: A new regulator of
osteoclast motility counteracting bone loss. J Bone Miner Res.
29:1833–1846. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Reppe S, Refvem H, Gautvik VT, Olstad OK,
Høvring PI, Reinholt FP, Holden M, Frigessi A, Jemtland R and
Gautvik KM: Eight genes are highly associated with BMD variation in
postmenopausal Caucasian women. Bone. 46:604–612. 2010. View Article : Google Scholar
|
|
7
|
Jemtland R, Holden M, Reppe S, Olstad OK,
Reinholt FP, Gautvik VT, Refvem H, Frigessi A, Houston B and
Gautvik KM: Molecular disease map of bone characterizing the
postmenopausal osteoporosis phenotype. J Bone Miner Res.
26:1793–1801. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Brazma A, Parkinson H, Sarkans U,
Shojatalab M, Vilo J, Abeygunawardena N, Holloway E, Kapushesky M,
Kemmeren P, Lara GG, et al: ArrayExpress-a public repository for
microarray gene expression data at the EBI. Nucleic Acids Res.
31:68–71. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bolstad BM, Irizarry RA, Astrand M and
Speed TP: A comparison of normalization methods for high density
oligonucleotide array data based on variance and bias.
Bioinformatics. 19:185–193. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Smyth GK: Limma: Linear models for
microarray data. Bioinformatics and computational biology solutions
using R and Bioconductor. Springer; pp. 397–420. 2005, View Article : Google Scholar
|
|
11
|
Benjamini Y and Hochberg Y: Controlling
the false discovery rate: A practical and powerful approach to
multiple testing. J R Stat Soc Series B Stat Methodol. 289–300.
1995.
|
|
12
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kanehisa M and Goto S: KEGG: Kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000. View Article : Google Scholar
|
|
14
|
Lachmann A, Xu H, Krishnan J, Berger SI,
Mazloom AR and Ma'ayan A: ChEA: Transcription factor regulation
inferred from integrating genome-wide ChIP-X experiments.
Bioinformatics. 26:2438–2444. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang J, Duncan D, Shi Z and Zhang B:
WEB-based GEne SeT AnaLysis Toolkit (WebGestalt): Update 2013.
Nucleic Acids Res. 41:4392013.
|
|
16
|
Szklarczyk D, Franceschini A, Kuhn M,
Simonovic M, Roth A, Minguez P, Doerks T, Stark M, Muller J, Bork
P, et al: The STRING database in 2011: functional interaction
networks of proteins, globally integrated and scored. Nucleic Acids
Res. 39:D561–D568. 2011. View Article : Google Scholar :
|
|
17
|
Smoot ME, Ono K, Ruscheinski J, Wang PL
and Ideker T: Cytoscape 2.8: new features for data integration and
network visualization. Bioinformatics. 27:431–432. 2011. View Article : Google Scholar :
|
|
18
|
Enright AJ, Van Dongen S and Ouzounis CA:
An efficient algorithm for large-scale detection of protein
families. Nucleic Acids Res. 30:1575–1584. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Morris JH, Apeltsin L, Newman AM, Baumbach
J, Wittkop T, Su G, Bader GD and Ferrin TE: ClusterMaker: A
multi-algorithm clustering plugin for Cytoscape. BMC
Bioinformatics. 12:4362011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Maere S, Heymans K and Kuiper M: BiNGO: A
Cytoscape plugin to assess overrepresentation of gene ontology
categories in biological networks. Bioinformatics. 21:3448–3449.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li B: Bone morphogenetic protein-Smad
pathway as drug targets for osteoporosis and cancer therapy. Endocr
Metab Immune Disord Drug Targets. 8:208–219. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
David L, Mallet C, Mazerbourg S, Feige JJ
and Bailly S: Identification of BMP9 and BMP10 as functional
activators of the orphan activin receptor-like kinase 1 (ALK1) in
endothelial cells. Blood. 109:1953–1961. 2007. View Article : Google Scholar
|
|
23
|
Li B: Bone morphogenetic protein-Smad
pathway as drug targets for osteoporosis and cancer therapy. Endocr
Metab Immune Disord Drug Targets. 8:208–219. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wu L, Wu Y, Gathings B, Wan M, Li X,
Grizzle W, Liu Z, Lu C, Mao Z and Cao X: Smad4 as a transcription
corepressor for estrogen receptor alpha. J Biol Chem.
278:15192–15200. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Genant HK, Lucas J, Weiss S, Akin M, Emkey
R, McNaney-Flint H, Downs R, Mortola J, Watts N, Yang HM, et al:
Low-dose esterified estrogen therapy: Effects on bone, plasma
estradiol concentrations, endometrium and lipid levels.
Estratab/Osteoporosis Study Group. Arch Intern Med. 157:2609–2615.
1997. View Article : Google Scholar
|
|
26
|
Cummings SR, Ensrud K, Delmas PD, LaCroix
AZ, Vukicevic S, Reid DM, Goldstein S, Sriram U, Lee A, Thompson J,
et al: Lasofoxifene in postmenopausal women with osteoporosis. N
Engl J Med. 362:686–696. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Caro JJ, Ishak KJ, Huybrechts KF, Raggio G
and Naujoks C: The impact of compliance with osteoporosis therapy
on fracture rates in actual practice. Osteoporos Int. 15:1003–1008.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Cheng P, Chen C, He HB, Hu R, Zhou HD, Xie
H, Zhu W, Dai RC, Wu XP, Liao EY, et al: miR-148a regulates
osteoclastogenesis by targeting V-maf musculoaponeurotic
fibrosarcoma oncogene homolog B. J Bone Miner Res. 28:1180–1190.
2013. View Article : Google Scholar
|
|
29
|
Wang Y, Li L, Moore BT, Peng XH, Fang X,
Lappe JM, Recker RR and Xiao P: MiR-133a in human circulating
monocytes: a potential biomarker associated with postmenopausal
osteoporosis. PLoS One. 7:e346412012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Cao Z, Moore BT, Wang Y, Peng XH, Lappe
JM, Recker RR and Xiao P: MiR-422a as a potential cellular microRNA
biomarker for postmenopausal osteoporosis. Plos One. 9:e970982014.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Seeliger C, Karpinski K, Haug AT, Vester
H, Schmitt A, Bauer JS and van Griensven M: Five freely circulating
miRNAs and bone tissue miRNAs are associated with osteoporotic
fractures. J Bone Miner Res. 29:1718–1728. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Di Vizio D, Freeman MR and Morello M:
Large oncosomes in human tumors and in circulation in patients with
cancer. U.S. Patent Application 13/975,059. 2013 8 23
|
|
33
|
Burgess DL, Gefrides LA, Foreman PJ and
Noebels JL: A cluster of three novel Ca2+ channel gamma subunit
genes on chromosome 19q13. 4: evolution and expression profile of
the gamma subunit gene family. Genomics. 71:339–350. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Giannini S, Nobile M, Dalle Carbonare L,
Lodetti MG, Sella S, Vittadello G, Minicuci N and Crepaldi G:
Hypercalciuria is a common and important finding in postmenopausal
women with osteoporosis. Eur J Endocrinol. 149:209–213. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Azuma K, Urano T, Ouchi Y and Inoue S:
Glucocorticoid-induced gene tripartite motif-containing 63 (TRIM63)
promotes differentiation of osteoblastic cells. Endocr J.
57:455–462. 2009. View Article : Google Scholar
|
|
36
|
Kondo H, Ezura Y, Nakamoto T, Hayata T,
Notomi T, Sorimachi H, Takeda S and Noda M: MURF1 deficiency
suppresses unloading-induced effects on osteoblasts and osteoclasts
to lead to bone loss. J Cell Biochem. 112:3525–3530. 2011.
View Article : Google Scholar : PubMed/NCBI
|