Introduction
In total, ~400,000,000 individuals are infected by
hepatitis B virus (HBV) worldwide, which is a leading risk factor
for hepatocellular carcinoma (HCC). During the process of HBV
infection, certain HBV DNA molecules may enter the nuclei and
integrate into the host chromosomal DNA, which is suspected to be
one of the major etiological events in HBV-induced HCC.
Conventional polymerase chain reaction (PCR)-based methods,
including Alu-PCR and inverse PCR, have technological limitations
in detecting the presence of viral integration, resulting in only a
small subset of insertions, or only the insertions close to the
targeted human or viral sequences being efficiently detected
(1–3). As a result, few HBV integration
breakpoints have been found through these methods, and these
findings may be of little oncogenic annotation.
With the rapid development of parallel sequencing
technology, whole genome sequencing (WGS) has provided novel
insight into HBV integration breakpoints in the HCC genome.
Recently, through the application of WGS, several studies have
reported a substantial number of unbiased and unprecedented HBV
integrations, and a few frequently targeted genes in HCC including
hTERT, MLL4 and CCNE1, have been identified simultaneously
(4,5). According to a previous study using
WGS (6), the major integration
site (MIS) (3), chr16: 51320015,
was identified, and the present study aimed to detect the presence
of this site in the hepatocytes of patients infected with chronic
hepatitis B (CHB). Furthermore, the present study aimed to examine
the significance of quantitative measurements of chr16:51320015 in
these patients.
Patients and methods
Patients and samples
In the present study, 30 hepatitis B e antigen
(HBeAg)-positive (+) and 30 HBeAg-negative (−) patients with CHB
were recruited from the Department of Infectious Diseases, Remin
Hospital, Hubei University of Medicine (Shiyan, China). CHB was
documented by the presence of HBV DNA in the serum for >6 months
and a serum alanine aminotransferase level greater than twice the
normal range (7). All patients
were treatment-naive. Patients who were co-infected with hepatitis
D, hepatitis C or human immunodeficiency virus, or those with
Wilson's disease, primary biliary cirrhosis or with a substantial
daily alcohol intake (20 g/day for females; 30 g/day for males)
were excluded from the investigation. Each patient signed an
informed consent document and the study was approved by the Ethics
Committee of Remin Hospital. Following collection, liver biopsy
specimens (~10 mg) were frozen in liquid nitrogen and stored at
−80°C, serum were stored at −30°C, respectively, until experimental
analysis.
IH HBV covalently closed circular DNA
(cccDNA) quantification
DNA was extracted from biopsy specimens using a
QIAamp® DNA Mini kit (Qiagen, Hilden, Germany). The
levels of intrahepatic (IH) covalently closed circular DNA (cccDNA)
were measured using reverse transcription-quantitative (RT-q)PCR
analysis, as described previously (8). β-globin DNA (housekeeping gene) was
detected using a LightCycler® Control kit DNA (Roche
Diagnostics GmbH, Mannheim, Germany) in order to count the cell
number in the biopsies and calculate the number of copies/cell.
Serum HBV DNA quantification
DNA was extracted from 200 µl serum using a
QIAamp® DNA Blood Mini kit (Qiagen), and serum HBV DNA
levels were measured using Cobas®TaqMan®
RT-qPCR, as described previously (Roche Diagnostics) (9).
Quantification of serum hepatitis B
surface antigen (HBsAg)
The levels of HBsAg were quantified using an enzyme
immunoassay with the Abbott ARCHITECT platform (Abbott
Laboratories, Abbott Park, IL, USA), according to the
manufacturer's instructions. HBsAg >0.05 IU/ml was considered to
indicate a positive result.
PCR and Sanger sequencing validation
Conventional PCR and Sanger sequencing were used to
verify the chr16: 51320015 integration site in the hepatocytes of
the patients. PCR primers for a 305 bp fragment were designed based
on WGS-assembled sequences, in which one primer located in human
genome and the other in HBV genome (forward
5′-GTCTTGCCCAAGGTCTTA-3′ and reverse 5′-CAGATGGCGCACTAACAA-3′). The
PCR mix was prepared as follows: 1 µl DNA; 2 µl
10×Taq Buffer; 11.5 µl H2O; 2.5 µl dNTPs;
1 µl forward and reverse primers (10 µM,
respectively); 1 µl hot start Taq™ enzyme (Takara Bio, Inc.,
Otsu, Japan). The following cycling conditions were used: Initial
denaturation for 30 sec at 95°C; 40 cycles of denaturation for 10
sec at 95°C, annealing for 10 sec at 56°C and extension for 14 sec
at 72°C, final extension for 7 min at 72°C. The PCR products were
electrophoresed through a 1% agarose gel, and then extracted and
sequenced using Sanger sequencing (Shanghai Sangon Biology
Engineering Technology and Service Co, Ltd., Shanghai, China).
Finally the results of the sequencing were compared with HBV and
the human genome using the Basic Local Alignment Search Tool
(BLAST; http://blast.ncbi.nlm.nih.gov/Blast.cgi#alnHdr).
Quantification of the chr16: 51320015
integration site
The PCR-amplified fragment of the chr16: 51320015
integration site was retrieved from the 1% agarose gels (Biowest
LLC, Kansas City, MO, USA) with 0.5 µg/ml ethidium bromide
(Promega Corporation, Madison, WI, USA) using a QIAquick Gel
Extraction kit (Qiagen) and inserted into a PMD 18-T vector (Takara
Bio, Inc.), which was electrotrans-formed into Escherichia
coli DH5α cells (Takara Bio, Inc.) successively, according to
the manufacturer's instructions. Following proliferation in
lysogeny broth culture medium containing 100 µg/ml
Ampicillin at 37°C for 16 h and blue-white screening, the fragment
containing the plasmid was extracted using a QIAfilter Plasmid Mini
kit (Qiagen) and quantified using nanodrop 2000 spectrophotometry
(Thermo Fisher Scientific, Waltham, MA, USA) at 260 nm. A series of
quantification standards were made by diluting the plasmid in
double distilled water. The standard dilutions were
5×107, 5×105, 5×104,
5×103 and 5×102 copies/cell. Consequently, a
20 µl reaction volume was used, containing 1 µl
extracted DNA, 0.8 µl of the above-mentioned forward and
reverse primers (10 µM), 7.4 µl nuclease-free water
and 10 µl 2X SYBR Green-I (Takara Bio, Inc.). SYBR Green I
RT-qPCR was performed using a LightCycler™ (Roche Diagnostics), and
the fluorescence was determined at 72°C. According to the
measurement of β-globin DNA, the numbers of chr16: 51320015
integration sites were detected and were compared as the number of
copies/cell.
Statistical analysis
Statistical analyses were performed using SPSS 13.0
statistical software (SPSS, Inc., Chicago, IL, USA). Continuous
variables are expressed as the mean ± standard error of the mean
and were analyzed using non-paired Student's t-tests. The levels of
serum HBsAg (IU/ml) and HBV DNA (copies/ml) were logarithmically
transformed prior to analysis. Categorical variables were compared
using Pearson's χ2 test. Correlations were analyzed
using Pearson's correlation coefficient. Two-sided P<0.05 was
considered to indicate a statistically significant difference.
Results
Baseline characteristics
The clinical, virological and serological
characteristics of the patient groups used in the present study are
listed in Table I. The HBeAg (+)
patients, comprising 26 males and four females) were aged between
12 and 59 years (35.4±7.4 years), and the HBeAg (−) patients (23
males and seven females) were aged between 13 and 51 years
(31.6±6.8 years; P>0.05). Serum HBV DNA levels were
significantly lower in the HBeAg (−) patients, compared with those
in the HBeAg (+) patients (P=0.001), and the serum levels of HBsAg
in the HBeAg (−) patients were lower than those in the HBeAg (+)
patients, although this was not a statistically significant
difference (P>0.05).
 | Table IClinical, virological and serological
parameters of HBeAg positive (+) patients and HBeAg negative (−)
patients. |
Table I
Clinical, virological and serological
parameters of HBeAg positive (+) patients and HBeAg negative (−)
patients.
| Parameter | HBeAg (+) | HBeAg (−) | P-value |
|---|
| Age (years) | 35.42 (12–59) | 31.64 (13–51) | 0.171 |
| Gender (M/F) | 26/4 | 23/7 | 0.317 |
| HBsAg
(log10 IU/ml) | 2.70
(−1.15–4.27) | 1.66 (−2–4.58) | 0.060 |
| HBV DNA
(log10 copies/ml) | 5.45 (2.71–8.13) | 4.10 (2.43–5.29) | 0.001 |
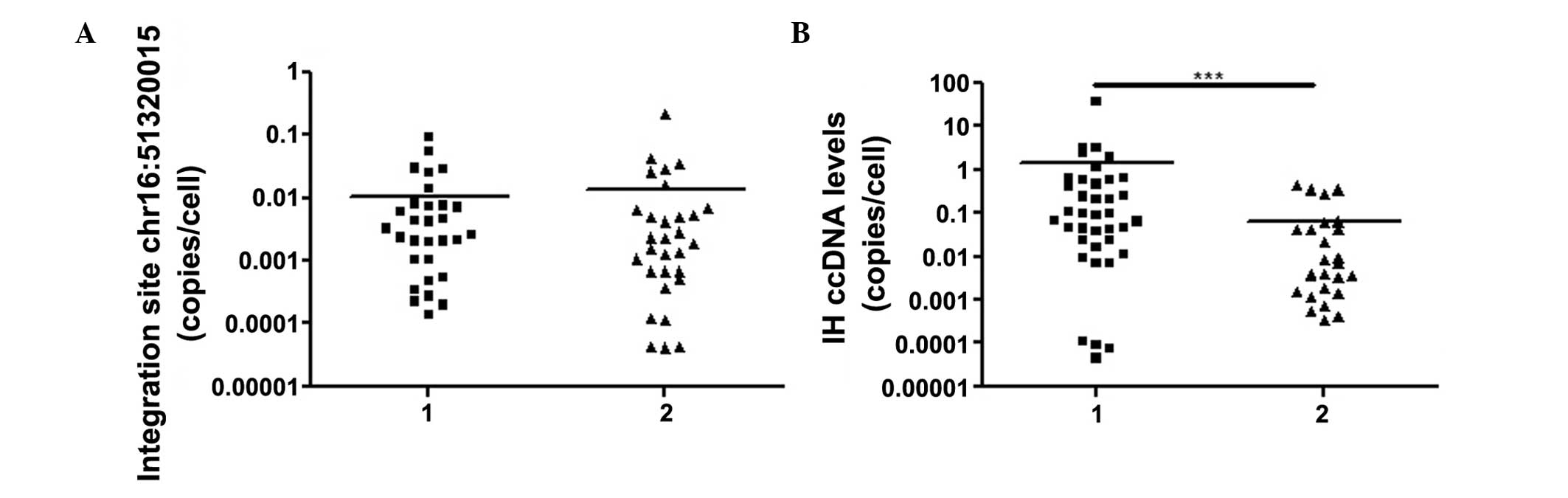
IH cccDNA quantification
The lower limit of detection for IH cccDNA was
2.4×10−4 copies/cell. The levels of IH cccDNA were
detectable in 26 of the HBeAg (−) patients and in all 30 of the
HBeAg (+) patients enrolled in the present study, and the number of
copies was significantly higher in the HBeAg (+) patients
(1.43±9.79×10−1 copies/cell), compared with the HBeAg
(−) patients (6.58×10−2±2.47×10−2
copies/cell; P<0.0001; Fig.
1).
Quantification of the chr16: 51320015
integration site
According to the results of the conventional RT-qPCR
and Sanger sequencing, the chr16: 51320015 integration site was
present in the hepatocytes of all the patients enrolled in the
presents study, the fragments of which were located in the
1,631–1,807 nt of the HBV sequence and the 51,320,015–51,319,900 nt
of the human sequence, respectively (Figs. 2 and 3). The average level of this site was
1.21×10−2±3.07×10−2 copies/cell
(4.16×10−5−0.212 copies/cell). No significant difference
was observed between The HBeAg (+) patients
(1.05×10−2±3.60×10−3 copies/cell) and the
HBeAg (−) patients (1.37×10−2±7.14×10−3
copies/cell; P>0.05; Fig.
1).
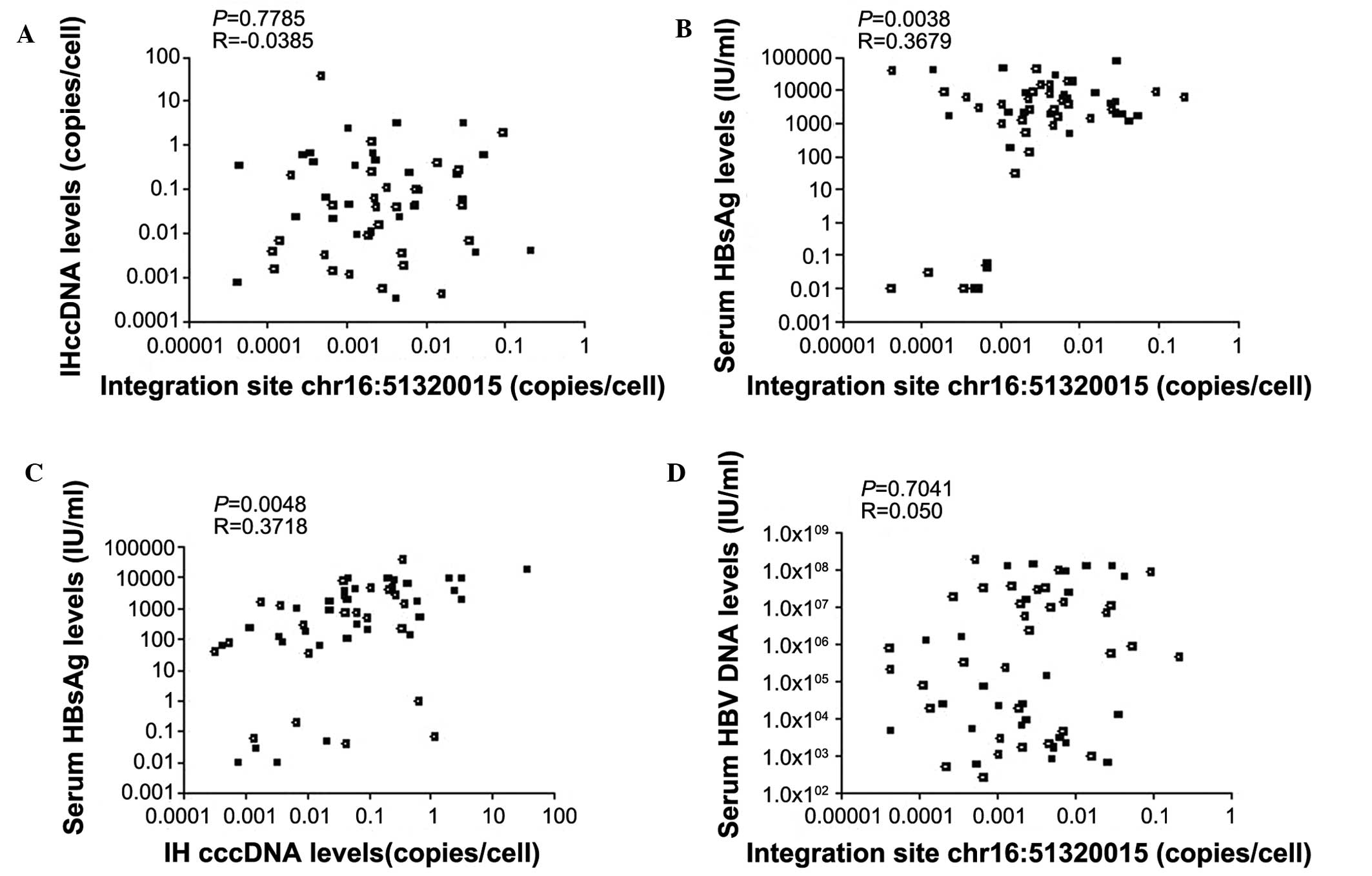
Correlation analysis
The number of copies of the chr16: 51320015
integration site were positively correlated with the serum levels
of HBsAg (P=0.0038), but not with the serum levels of HBV DNA or IH
cccDNA (P=0.7041 and P=0.7785, respectively). A weak correlation
was observed between the levels of IH cccDNA and the serum levels
of HBsAg (P=0.0048; Fig. 4).
Discussion
Following the process of early or persistent HBV
infection, relaxed-circle DNA (rcDNA) is transferred to the nucleus
of hepatocytes, where it forms cccDNA, the virus transcriptional
template (10,11). Within infected cells, pregenomic
RNA and is then transcribed from the cccDNA and is transported to
the cytoplasm, where the mature capsids of the rcDNA are reverse
transcribed and either secreted from the cells or returned to the
nucleus to form the cccDNA pool. During the formation of cccDNA,
linear HBV DNA, including double linear DNA and single-stranded
DNA, produced through illegitimate replication and deficient HBV
transcription, may integrate into the host chromosomal DNA
(12). According to previous
studies involving the application of WGS (13,14),
at least two promulgated mechanisms may be involved in the
oncogenicity of HBV integration: (i) HBV DNA insertion into the
host genome altering the function of endogenous genes, and inducing
chromosomal instability and changes in copy numbers; (ii)
expression of C-terminal truncated HBx or HBs protein, which may
modulate cell proliferation and viability. In addition, the immune
response against virally-infected cells may be induced by the
protein transcribed from integrated DNA, for example, the x gene.
Thus, it is very important to effectively recognize and eradicate
hepatocytes with integrated DNA in the treatment of CHB (6).
Early reports stated that HBV integration events may
be randomly distributed across the whole genome (2,3).
However, increasing evidence has indicated that several genes may
be preferentially integrated by the viral DNA, for example,
chromosomes 10 and 17 (5,6). The present study demonstrated that
chr16: 51320015 may also be favorably integrated, and this junction
was found to occrr in the hepatocytes of all the patients with CHB
enrolled in the present study. The junction of its inserted viral
fragment was at 1,807 nt, within the DR2-DR1 region of the HBV
genome (1,590–1,834 nt). The DR2 and DR1 sites represent the ends
of the partially duplex HBV DNA and can provide DNA termini for
non-homologous end joining (NHEJ). Consequently these sites are
more likely to be the initiation break points for HBV integration
(15). However, in human chromatin
HBV, integration events are more likely to occur in regions which
are characterized by either looser secondary structures or open
chromatin configuration, which facilitate breakage and provide DNA
termini for NHEJ with HBV DNA (16). At present, it is difficult to
recognize and eradicate hepatocytes with viral integration of
patients with CHB, however, the above findings may provide a novel
perspective on either the diagnostic or therapeutic strategies of
HBV integration, for example, chimeric antigen receptor therapy
(17–19).
The liver is a closed, self-renewing population of
cells, in which hepatocytes are generally long lived with a
lifetime reported to exceed 6 months (20). During chronic HBV infection,
hepatocyte transformation usually results from mutations that are
caused by persistent inflammation, leading to cumulative oxidative
damage to the host DNA (21). In
addition, this environment provides the opportunity for the
expansion of hepatocyte variants with a selective advantage, either
in growth or survival (22).
Although the basis of the clonal expansion in the hepatocytes
remains to be fully elucidated, there are several explanations
(23). A possible explanation
involves cellular transformation leading to unregulated growth,
however, this does not explain clones with no clear morphological
transformation. A second explanation involves random death and
regeneration within the entire hepatocyte population, however, is
unlikely to explain the occurrence of very large clones of
>104 hepatocytes. Another explanation for large
clones of hepatocytes involves the resident stem/progenitor cells,
however, this is not supported by the current knowledge (23). Finally, a model in which immune
evasion caused by HBV integration is the basis for clonal
expansions is favored, as although smaller cell clones may be the
result of random turnover, the presence of copy clones of
>105 cells requires alternative explanations
(24).
Although HBV integration occurs at random sites in
host DNA, and each integration event provides a unique genetic
marker for the cells in which it occurred, the unique viral-cell
junctions of integrated DNA may be used to track clonal
proliferation of hepatocytes (22). Traditionally, clonal expansion was
detected by assaying for integrated HBV DNA using inverse PCR.
However, the level of clonal expansion may be underestimated using
this technique, which is only suitable for detecting the
integration of viral DNA close to particular restriction
endonuclease cleavage sites in host DNA, and not all integrations
can be detected using only a single enzyme (24). Thus, the present study aimed to
investigate the clonal expansion of hepatocytes using quantitative
measurements of the chr16: 51320015 integration site. The average
level of this site was determined to be
1.21×10−2±3.07×10−2 copies/cell
(4.16×10−5−0.212 copies/cell), indicating that this
integration site may have originated from clonal expansion, while
high-copy clones with detectable integrated DNA have been estimated
at a frequency of >2×10−6 copies/cell (22).
According to quantitative measurements of serum
HBeAg, patients with CHB can be divided into HBeAg (+) patients and
HBeAg (−) patients, and HBeAg seroconversion and loss usually
signify that HBV replication has been effectively suppressed by the
host immunity (25). However, in
the present study, although the levels of IH cccDNA and serum HBV
DNA were significantly higher in the HBeAg (+) patients, compared
with those in the HBeAg (−) patients, no significant differences in
the number of copies of the chr16: 51320015 integration site and
serum levels of HBsAg were found between the two patient groups.
The copy numbers of chr16: 51320015 integration site were
positively correlated with serum levels of HBsAg, but not with the
levels of IH cccDNA. These findings may be due to the different
origins of IH cccDNA and HBV integration. While the accumulation of
IH cccDNA may be due to rcDNA recurrently entering into the
nucleus, those of the chr16: 51320015 integration site, as
described above, may have originated from the clonal expansion of
integrated hepatocytes. Furthermore, the production of HBsAg in the
HBeAg (−) patients, which is independent of HBV replication, was
abundant and far exceeded that required for virion assembly. This
may be partially produced from HBV integration (26–28),
explaining the significant association between the copy numbers of
the chr16: 51320015 integration site and serum levels of HBsAg, but
weak association with the levels of IH cccDNA. Consequently, the
present study hypothesized that, in patients with CHB, the
accumulation of HBV integration may not be effectively suppressed,
even when the production of HBV is completely controlled by host
immunity. These findings are in agreement with the hypothesis that,
in addition to severe liver damage, HBV integration may also be a
prerequisite for HCC (29).
In conclusion, the present study demonstrated that
the chr16: 51320015 integration site was present in the hepatocytes
of all the patients with CHB, which may have accumulated according
to clonal expansion. In addition, the number of copies of this site
were positively correlated with the serum levels of HBsAg, but not
with the levels of IH cccDNA. Whether this integration site occurs
in the hepatocytes of all patients infected with HBV requires
further investigation, as does its mechanism. In addition, whether
or not hepatocytes with HBV integration can be effectively
recognized and eradicated by means of this integration site
requires elucidation.
Acknowledgments
This study was supported by grants from the National
Natural Science Foundation of Hubei Province (grant no.
2015CFB290).
References
|
1
|
Murakami Y, Saigo K, Takashima H, Minami
M, Okanoue T, Bréchot C and Paterlini-Bréchot P: Large scaled
analysis of hepatitis B virus (HBV) DNA integration in HBV related
hepatocellular carcinomas. Gut. 54:1162–1168. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Saigo K, Yoshida K, Ikeda R, Sakamoto Y,
Murakami Y, Urashima T, Asano T, Kenmochi T and Inoue I:
Integration of hepatitis B virus DNA into the myeloidl/ymphoid or
mixed-lineage leukemia (MLL4) gene and rearrangements of MLL4 in
human hepatocellular carcinoma. Hum Mutat. 29:703–708. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Murakami Y, Minami M, Daimon Y and Okanoue
T: Hepatitis B virus DNA in liver, serum and peripheral blood
mononuclear cells after the clearance of serum hepatitis B virus
surface antigen. J Med Virol. 72:203–214. 2004. View Article : Google Scholar
|
|
4
|
Jiang Z, Jhunjhunwala S, Liu J, Haverty
PM, Kennemer MI, Guan Y, Lee W, Carnevali P, Stinson J, Johnson S,
et al: The effects of hepatitis B virus integration into the
genomes of hepa-tocellular carcinoma patients. Genome Res.
22:593–601. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sung WK, Zheng H, Li S, Chen R, Liu X, Li
Y, Lee NP, Lee WH, Ariyaratne PN, Tennakoon C, et al: Genome-wide
survey of recurrent HBV integration in hepatocellular carcinoma.
Nat Genet. 44:765–769. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Toh ST, Jin Y, Liu L, Wang J, Babrzadeh F,
Gharizadeh B, Ronaghi M, Toh HC, Chow PK, Chung AY, et al: Deep
sequencing of the hepatitis B virus in hepatocellular carcinoma
patients reveals enriched integration events, structural
alterations and sequence variations. Carcinogenesis. 34:787–798.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chinese Society of Hepatology and Chinese
Society of Infectious Diseases; Chinese Medical Association: The
guideline of prevention and treatment for chronic hepatitis B (2010
version). Zhonghua Gan Zang Bing Za Zhi. 19:13–24. 2011.In
Chinese.
|
|
8
|
Bowden S, Jackson K, Littlejohn M and
Locarnini S: Quantification of HBV covalently closed circular DNA
from liver tissue by real-time PCR. Methods Mol Med. 95:41–50.
2004.PubMed/NCBI
|
|
9
|
Weinberger KM, Wiedenmann E, Böhm S and
Jilg W: Sensitive and accurate quantitation of hepatitis B virus
DNA using a kinetic fluorescence detection system (TaqMan PCR). J
Virol Methods. 85:75–82. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ruan P, Zhou B, Dai X, Sun Z, Guo X, Huang
J and Gong Z: Predictive value of intrahepatic hepatitis B virus
covalently closed circular DNA and total DNA in patients with acute
hepatitis B and patients with chronic hepatitis B receiving
anti-viral treatment. Mol Med Rep. 9:1135–1141. 2014.PubMed/NCBI
|
|
11
|
Belloni L, Allweiss L, Guerrieri F,
Pediconi N, Volz T, Pollicino T, Petersen J, Raimondo G, Dandri M
and Levrero M: IFN-α inhibits HBV transcription and replication in
cell culture and in humanized mice by targeting the epigenetic
regulation of the nuclear cccDNA minichromosome. J Clin Invest.
122:529–537. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chou YC, Jeng KS, Chen ML, Liu HH, Liu TL,
Chen YL, Liu YC, Hu CP and Chang C: Evaluation of transcriptional
efficiency of hepatitis B virus covalently closed circular DNA by
reverse transcription-PCR combined with the restriction enzyme
digestion method. J Virol. 79:1813–1823. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Fujimoto A, Totoki Y, Abe T, Boroevich KA,
Hosoda F, Nguyen HH, Aoki M, Hosono N, Kubo M, Miya F, et al:
Whole-genome sequencing of liver cancers identifies etiological
influences on mutation patterns and recurrent mutations in
chromatin regulators. Nat Genet. 44:760–764. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li WY, Zeng X, Lee NP, Liu X, Chen S, Guo
B, Yi S, Zhuang X, Chen F, Wang G, et al: HIVID: An efficient
method to detect HBV integration using low coverage sequencing.
Genomics. 102:338–344. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li X, Zhang J, Yang Z, Kang J, Jiang S,
Zhang T, Chen T, Li M, Lv Q, Chen X, et al: The function of
targeted host genes determines the oncogenicity of HBV integration
in hepatocellular carcinoma. J Hepatol. 60:975–984. 2014.
View Article : Google Scholar
|
|
16
|
Ding D, Lou X, Hua D, Yu W, Li L, Wang J,
Gao F, Zhao N, Ren G, Li L and Lin B: Recurrent targeted genes of
hepatitis B virus in the liver cancer genomes identified by a
next-generation sequencing-based approach. PLoS Genet.
8:e10030652012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Bohne F, Chmielewski M, Ebert G, Wiegmann
K, Kürschner T, Schulze A, Urban S, Krönke M, Abken H and Protzer
U: T cells redirected against hepatitis B virus surface proteins
eliminate infected hepatocytes. Gastroenterology. 134:239–247.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Krebs K, Böttinger N, Huang LR,
Chmielewski M, Arzberger S, Gasteiger G, Jäger C, Schmitt E, Bohne
F, Aichler M, et al: T cells expressing a chimeric antigen receptor
that binds hepatitis B virus envelope proteins control virus
replication in mice. Gastroenterology. 145:456–465. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Díaz-Montero CM, Naga O, Zidan AA, Salem
ML, Pallin M, Parmigiani A, Walker G, Wieder E, Komanduri K, Cole
DJ, et al: Synergy of brief activation of CD8 T-cells in the
presence of IL-12 and adoptive transfer into lymphopenic hosts
promotes tumor clearance and anti-tumor memory. Am J Cancer Res.
1:882–896. 2011.PubMed/NCBI
|
|
20
|
Vemuru RP, Aragona E and Gupta S: Analysis
of hepatocellular proliferation: study of archival liver tissue is
facilitated by an endogenous marker of DNA replication. Hepatology.
16:968–973. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hagen TM, Huang S, Curnutte J, Fowler P,
Martinez V, Wehr CM, Ames BN and Chisari FV: Extensive oxidative
DNA damage in hepatocytes of transgenic mice with chronic active
hepatitis destined to develop hepatocellular carcinoma. Proc Natl
Acad Sci USA. 91:12808–12812. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Mason WS, Jilbert AR and Summers J: Clonal
expansion of hepatocytes during chronic woodchuck hepatitis virus
infection. Proc Natl Acad Sci USA. 102:1139–1144. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Mason WS, Liu C, Aldrich CE, Litwin S and
Yeh MM: Clonal expansion of normal-appearing human hepatocytes
during chronic hepatitis B virus infection. J Virol. 84:8308–8315.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Mason WS, Low CL, Xu C, Aldrich CE,
Scougall CA, Grosse A, Clouston A, Chavez D, Litwin S, Peri S, et
al: Detection of clonally expanded hepatocytes in chimpanzees with
chronic hepatitis B virus infection. J Virol. 83:8396–8408. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Volz T, Lutgehetmann M, Wachtler P, Jacob
A, Quaas A, Murray JM, Dandri M and Petersen J: Impaired
intrahepatic hepatitis B virus productivity contributes to low
viremia in most HBeAg-negative patients. Gastroenterology.
133:843–852. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Nguyen T, Thompson AJ, Bowden S, Croagh C,
Bell S, Desmond PV, Levy M and Locarnini SA: Hepatitis B surface
antigen levels during the natural history of chronic hepatitis B: A
perspective on Asia. J Hepatol. 52:508–513. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Thompson AJ, Nguyen T, Iser D, Ayres A,
Jackson K, Littlejohn M, Slavin J, Bowden S, Gane EJ, Abbott W, et
al: Serum hepatitis B surface antigen and hepatitis B e antigen
titers: Disease phase influences correlation with viral load and
intrahepatic hepatitis B virus markers. Hepatology. 51:1933–1944.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Manesis EK, Papatheodoridis GV, Tiniakos
DG, Hadziyannis ES, Agelopoulou OP, Syminelaki T, Papaioannou C,
Nastos T and Karayiannis P: Hepatitis B surface antigen: Relation
to hepatitis B replication parameters in HBeAg-negative chronic
hepatitis B. J Hepatol. 55:61–68. 2011. View Article : Google Scholar
|
|
29
|
Bonilla Guerrero R and Roberts LR: The
role of hepatitis B virus integtation in the pathogenesis of human
hepatocellular carcinoma. J Hepatol. 42:760–777. 2005. View Article : Google Scholar : PubMed/NCBI
|