Introduction
Proteomics is becoming increasingly important for
the development of novel biomarkers of various diseases (1). Serum contains a large variety of
proteins (2), which could reflect
the health of an individual or the current state of a patient
(3). Additionally, the collection
of serum is minimally invasive (4). Given this, a rich array of techniques
have been used to detect biomarkers of disease in serum.
Disease biomarkers for diagnostic and prognostic
purposes are most likely within an extremely low concentration
range (5). However, high-abundance
proteins in serum, including albumin and immunoglobulin G (IgG),
tend to mask low-abundance proteins. Thus, the depletion of albumin
and IgG is necessary for the detection of biomarkers (1,6).
At present, various techniques and methods have been
rapidly developed to deplete albumin and IgG (7). There are three main techniques,
including centrifugal ultrafiltration (size-based),
immunoaffinity-mediated proteomic separation (antibody-based) and
Cibacron Blue and related dye (affinity-based). Centrifugal
ultrafiltration is a cheap, convenient method but is not as
effective and has a lower reproducibility. Immunoaffinity-mediated
proteomic separation has a higher specificity, but is expensive and
fails to remove the associated fragments of target proteins. In
addition, this technique has a lower sample capacity. Furthermore,
it occasionally requires specific techniques, including
high-performance liquid chromatography. Dye-ligand affinity
chromatography is cheap, has a higher loading capacity and avoids
cross-contamination between different samples (8). However, the disadvantage of this
technique is a lack of specificity.
Based on the analysis of these methods, in the
present study, the Cibacron Blue columns were selected for further
assessment. The ProteoPrep Blue Albumin and IgG Depletion kit
(PROTBA; Sigma-Aldrich, St. Louis, MO, USA) and the Albumin and IgG
Removal kit (GE Healthcare, Waukesha, WI, USA) are two commonly
used commercial kits. However, to the best of our knowledge, no
study has yet compared the efficiencies of these two kits. In
addition, few studies have focused on how to optimize suitable
isoelectric focusing (IEF) protocols for solutions from the above
two kits. IEF is a critical step of two-dimensional electrophoresis
(2-DE). The solution obtained from the depletion of highly abundant
proteins contains certain materials, including ions, which
interfere with downstream IEF. Therefore, the aim of the present
study was to evaluate two commercial kits with regards to depletion
efficiency, specificity and capacity. In addition, the present
study also aimed to optimize the suitable downstream IEF protocol
and assess the reproducibility of this approach using the serum of
different patients.
Materials and methods
Materials
Strips, urea, thiourea, dithiothreitol (DTT),
immobilized pH gradient (IPG) buffer, CHAPS, iodoacetamide,
glycine, SDS and G250 were all purchased from GE Healthcare.
Acetone, methanol, ethanol and acetonitrile were obtained from
Sigma-Aldrich. Deionized water was prepared using the Milli-Q
system (Millipore Corporation, Bedford, MA, USA) and used for the
preparation of all buffers. Ettan DALTsix, Ettan IPGphor 3, Image
Scanner II and ImagerMaster software, version 7.0 were purchased
from GE Healthcare. The experiments of the current study were
approved by the ethics committee of the Guangzhou Hospital of
Traditional Chinese Medicine (Guangzhou, China). Written informed
consent was obtained from the patients prior to participation.
Serum samples
Serum obtained from two patients with colon cancer
(age, 40–64 years; male:female, 1:1) was separated by
centrifugation at 3,000 × g for 15 min at 4°C. The supernatant
aliquot was placed into tubes (500 µl per tube) and stored
at −80°C until use.
Removal of high-abundance proteins from
serum
Albumin and IgG were depleted according to the
manufacturer's instructions. Subsequently, protein quantification
was performed using the Quick Start Bradford Protein Assay (Bio-Rad
Laboratories, Inc., Hercules, CA, USA) and absorbance was measured
at 595 nm using the Multiskan Spectrum microplate reader (Bio-Rad
Laboratories, Inc.), using bovine serum albumin as a protein
standard.
SDS-PAGE
One-dimensional-SDS-PAGE
The depletion efficiency of each approach was
evaluated using 10% polyacrylamide gel. The sample was mixed with
2X sample buffer (GE Healthcare), loaded into each lane and
electrophoresed through a Bio-Rad system (Bio-Rad Laboratories,
Inc.) using the Laemmli SDS buffering system (25 mM Tris-base, 192
mM glycine and 0.1% SDS; Bio-Rad Laboratories, Inc.). The gels were
stained with Coomassie Blue (GE Healthcare) overnight according to
the manufacturer's instructions.
First-dimension separation
Following high-abundance protein depletion, 750
µg of protein and rehydration buffer was applied to the 18
cm ReadyStrip™ IPG strips (pH 4–7; GE Healthcare). IEF was
performed using an IPG III system (GE Healthcare). All the
processes above were performed at 20°C. The focused IEF strips were
stored at −80°C.
Two-dimensional separation
IPG strips were equilibrated for 15 min in 50 mM
Tris-HCl (pH 8.8), containing 6 M urea, 1% w/v SDS, 30% v/v
glycerol and 65 mM DTT (GE Healthcare), and re-equilibrated for 15
min in the same buffer containing 260 mM iodoacetamide in place of
DTT. Two-dimensional gel electrophoresis was performed on 12.5%
SDS-PAGE gel, using the Ettan DALTsix system (GE Healthcare).
Two-dimensional gels were run at 1 w per gel for 1 h at 15°C and
then 15 w per gel until the tracking dye migrated to within 1 cm of
the bottom of the gel.
Gel staining
The gels were washed in distilled water and stained
with colloidal Coomassie or silver (GE Healthcare) according to the
manufacturer's instructions.
Image analysis
The 2-DE gel was transferred into distilled water
and scanned using the ImageScanner system (GE Healthcare) combined
with LabScan software, version 7.0 (GE Healthcare). All gel images
were analyzed using ImageMaster 2D Platinum software (GE
Healthcare). Gel images from each group were edited and spots were
matched. A unique identification number was assigned to matching
spots on different gels. Normalization of the spot intensities was
conducted according to the total optical density of the gel.
Matrix-assisted laser
desorption/ionization mass spectrometry (MALDI-MS) and MS/MS
analysis
MALDI-time of flight (TOF)/TOF MS measurements were
performed on a Bruker Ultraflex III MALDI-TOF/TOF mass Spectrometer
(Bruker Daltonics, Leipzig, Germany) operating in reflectron mode
with a 20 kV accelerating voltage and 23 kV reflecting voltage. A
saturated solution of α-cyano-4-hydroxycinnamic acid in 50%
acetonitrile and 0.1% trifluoroacetic acid was used as the matrix.
The matrix solution (1µl) and sample solution at a ratio of
1:1 was applied onto the Score384 target well. A standard peptide
calibration mix in the mass range 800–3,200 Da (Bruker Daltonics)
was analyzed for external calibration of the mass spectrometer. The
calibration mix contained: Angiotensin II, angiotensin I, substance
P, bombesin, ACTH clip 1–17, ACTH clip 18–39 and somatostatin 28. A
series of eight samples were spotted around one external
calibration mixture. The SNAP algorithm (S/N threshold: 5; Quality
Factor Threshold: 30) in Flex Analysis 3.0 (Bruker Daltonics) was
used to identify the 100 most prominent peaks in the mass range m/z
700–4,000. The subsequent MS/MS analysis was performed in a
data-dependent manner and the five most abundant ions fulfilling
certain preset criteria (S/N higher than 3 and Quality Factor
higher than 30) were subjected to high energy collision-induced
dissociation analysis. The collision energy was set to 1 keV and
nitrogen was used as the collision gas.
Database searching
Peptide mass fingerprints (PMFs) were searched using
the program Mascot 2.1 (Matrix Science Ltd., London, UK) against
the SwissProt database (version 20091028, 510076 sequences;
http://www.uniprot.org/uniprot). The
search parameters were as follows: Trypsin digestion with one
missed cleavage; carbamidomethyl modification of cysteine as a
fixed modification and oxidation of methionine as a variable
modification; peptide tolerance maximum, ±0.3 Da; MS/MS tolerance
maximum, ±50 ppm; peptide charge, +1; monoisotopic mass. P<0.05
was for a local PMF search. For unambiguous identification of
proteins, more than five peptides must be matched for a PMF
search.
Different protocols for IEF
The different protocols for IEF were as follows:
Protocol A, 1 h at 200 v, 2 h at 10 kv and a final voltage of 8 kv
to 80 k vhs; protocol B, 2 h at 200 v and a final voltage of 8 kv
to 80 k vhs; protocol C, 10 h at 30 v and a final voltage of 10 kv
to 100 k vhs; protocol D, 3 h at 200 v and a final voltage of 10 kv
to 110 k vhs; protocol E, final voltage of 10 kv to 130 k vhs.
Results
Depletion efficiency of the kits
The ProteoPrep Blue Albumin and IgG Depletion kit
and Albumin and IgG Removal kit are commonly used commercial kits
to deplete albumin and IgG. Therefore, in the present study these
two kits were compared with regards to depletion efficiency,
specificity and capacity.
The present study initially used SDS-PAGE to compare
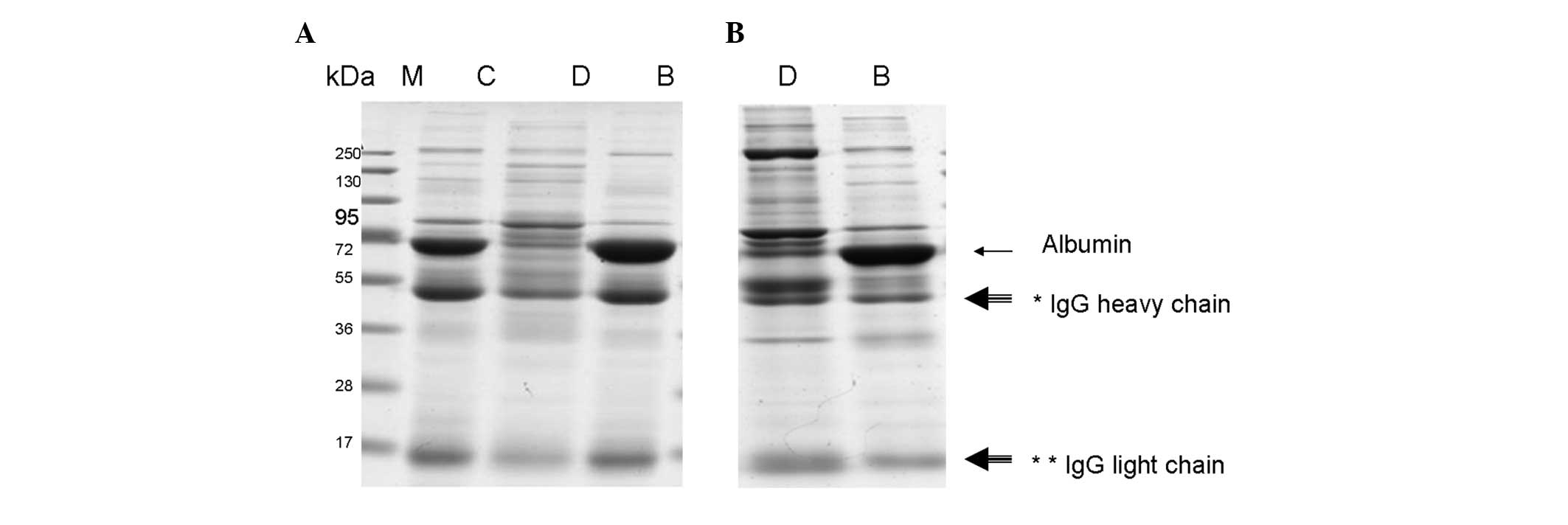
the efficiency of these two kits. As shown in Fig. 1A, albumin and IgG were almost
completely removed by the PROTBA-Sigma kit (Fig. 1A, Lane B). Following depletion,
albumin and IgG bound to the column (Fig. 1A, Lane C). By contrast, the Albumin
and IgG Removal kit could not effectively deplete albumin and IgG
(Fig. 1B, Lane B), and a
considerable quantity remained in the serum (Fig. 1B, Lane D). This result suggests
that the ProteoPrep Blue Albumin and IgG Depletion kit could
effectively deplete albumin and IgG.
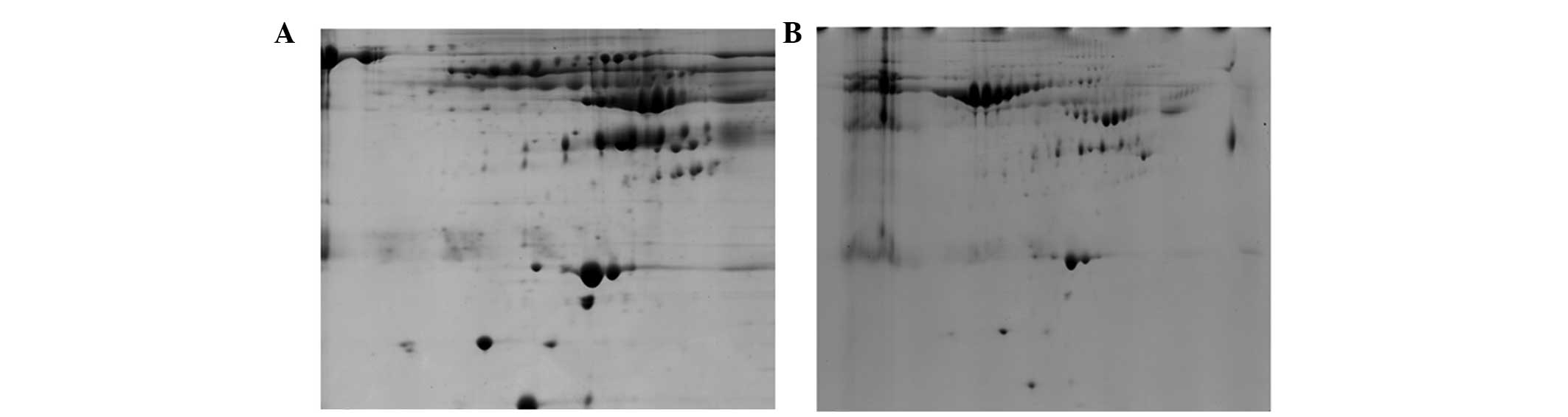
The present study aimed to investigate the depletion
effect of the above kits. The samples obtained by these two kits
were further separated by 2-DE. As shown in Fig. 2, the number of protein spots on the
2D gel obtained from the ProteoPrep Blue Albumin and IgG Depletion
kit was higher than that observed from the Albumin and IgG Removal
kit (540±43 vs. 400±23, respectively). The above results
demonstrated that the PROTBA-Sigma kit had a higher efficiency
compared with the Albumin and IgG removal kit.
Loading capacity/recovery of protein
The protein yield post-deletion is important for
downstream analysis. Therefore, the post-deletion protein yield of
these two kits was compared. As shown in Table I, compared with the Albumin and IgG
Removal kit, the ProteoPrep Blue Albumin and IgG Depletion kit
could load a higher volume (25–50 µl vs. 15–30 µl)
and produce a higher protein yield (260–540 µg vs. 150–250
µg). Notably, the solution obtained from the ProteoPrep Blue
Albumin and IgG Depletion kit can be applied to IEF directly.
 | Table IApproximate yield from two kits. |
Table I
Approximate yield from two kits.
| Kit name | Loading volume
(µl) | Loading capacity
(µg) | Recovery protein | Yield (%) | Directly apply to
IEF |
|---|
| ProteoPrep Blue
Albumin and IgG Depletion kit | 25–50 | 1,500–3,000 | 260 µg/25
µl
540 µg/50 µl | 17.33–18.00 | Yes |
| Albumin and IgG
removal kit | 15–30 | 900–1,800 | 150 µg/15
µl
250 µg/30 µl | 13.66–13.80 | No |
Specificity and protein
identification
Since the above results demonstrated that the
ProteoPrep Blue Albumin and IgG Depletion kit had a higher capacity
and higher depletion efficiency, this kit was selected to further
assess its specificity.
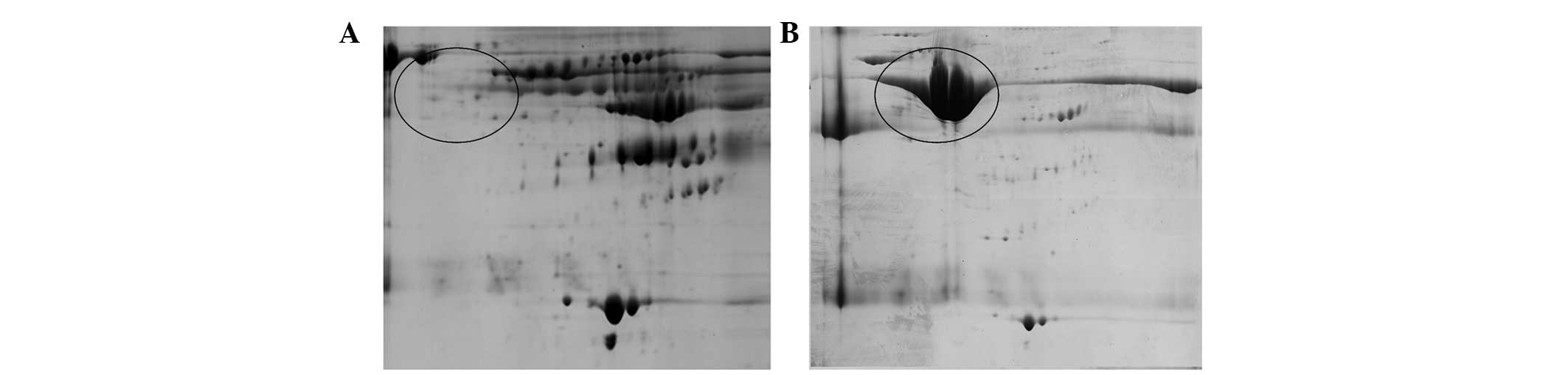
2D gel was used to analyze samples obtained from the
depletion of highly abundant proteins. As shown in Fig. 3, this kit depleted almost all
albumin and IgG. By contrast, 2D gel of eluate had more albumin
(square) and IgG (ellipse) and less low abundance protein.
Optimization of the IEF protocol
Following the depletion of highly abundant proteins,
serum contains additional ions, which may affect downstream IEF. In
order to obtain a suitable downstream IEF protocol, five different
IEF protocols were assessed as described in Materials and methods.
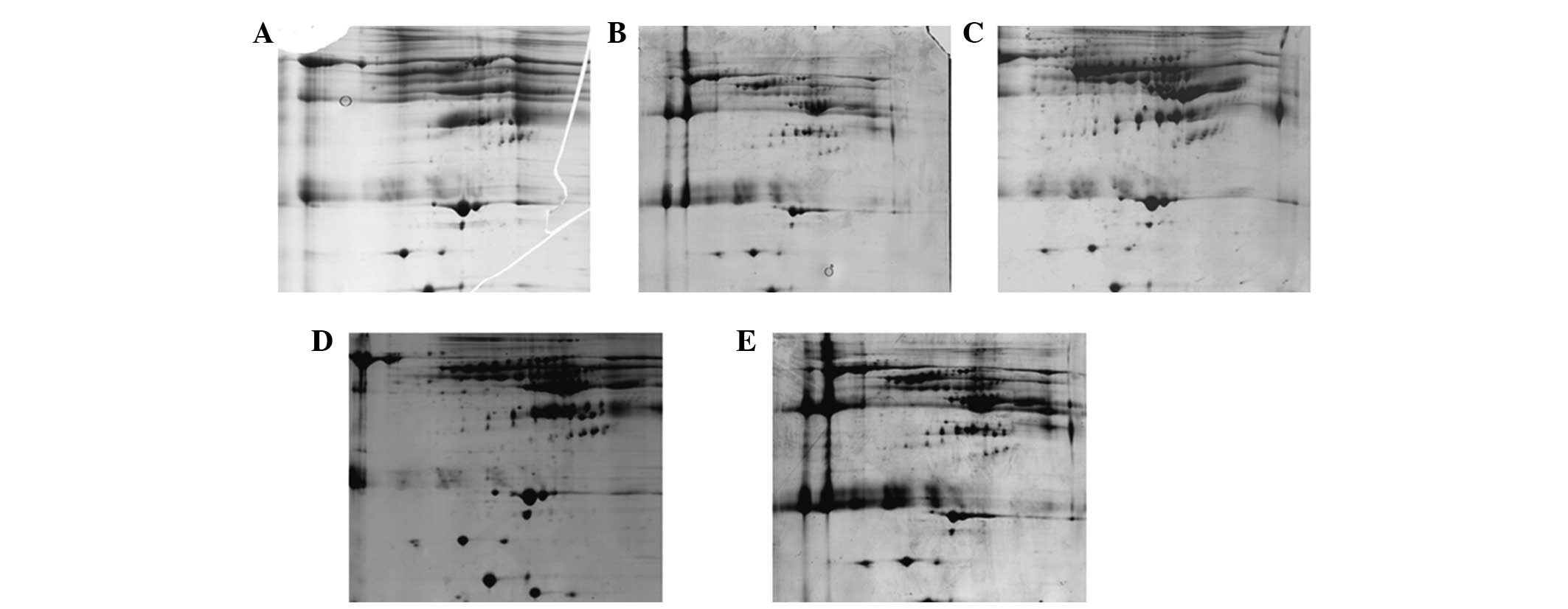
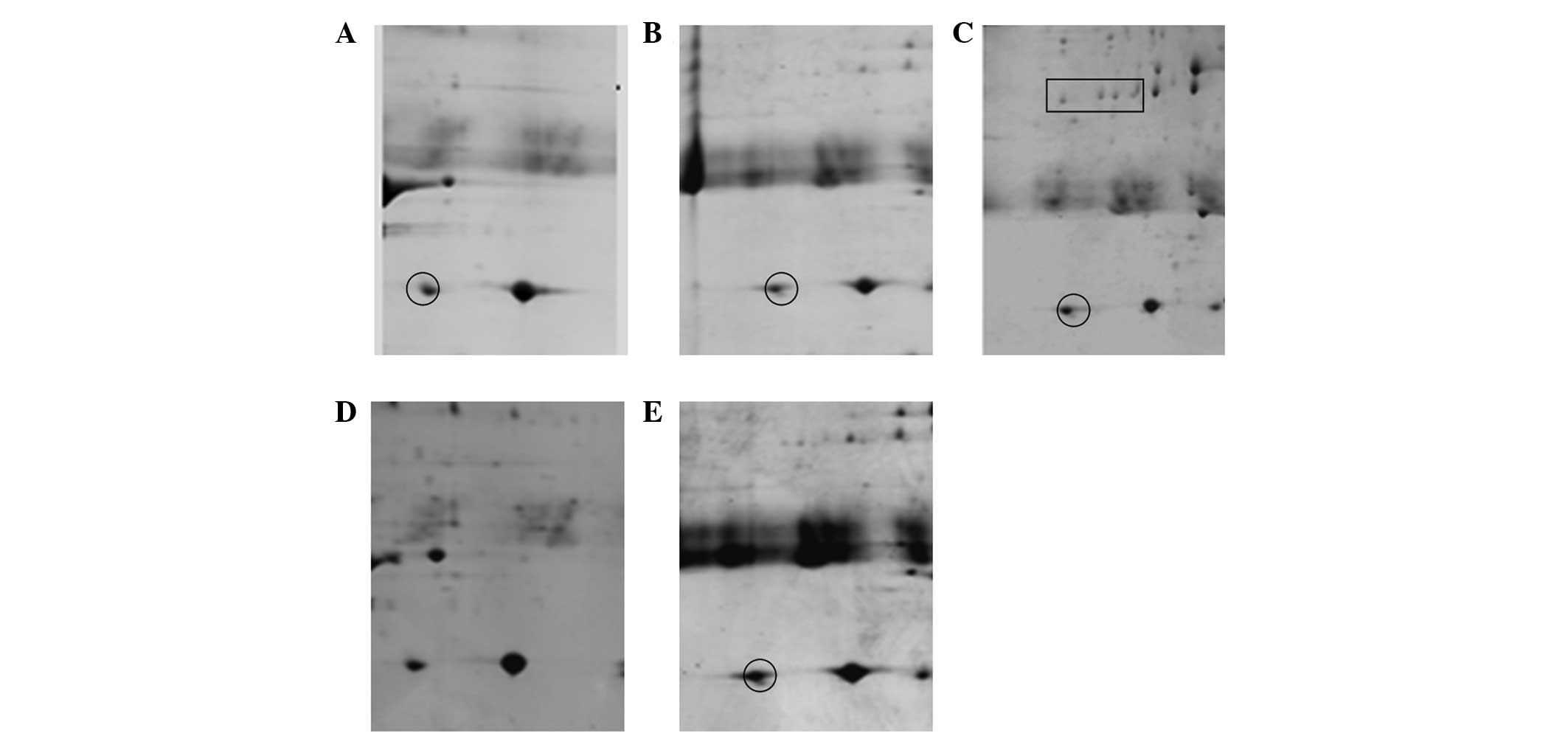
Fig. 4 shows that Protocol A and B
led to fewer spots and numerous streaks (Fig. 4A and B). This indicated that
proteins form aggregates during IEF. The optimal protocol was C,
which resulted in a good resolution of protein spots, less
horizontal streaks and the greatest number of spots (Fig. 4C). Protocol D and E produced more
spots but also produced more horizontal streaks (Fig. 4D and E). The number of protein
spots on 2-D gels of protocol A, B, C, D and E was 43±8, 134±18,
538±45, 443±32 and 404±33, respectively.
Subsequently, five unique spots (Fig. 5) from the depleted gels were
randomly selected for identification by MALDI-TOF MS. As shown in
Table II, the majority of the
protein spots, including apolipoprotein E, serotransferrin, heat
shock 70 kDa protein 1, tubulin α chain-like 3 and tropomyosin β
chain, were predominantly low-abundance proteins.
 | Table IIProteins identified from image C,
identified by matrix-assisted laser desorption ionization
time-of-flight MS. |
Table II
Proteins identified from image C,
identified by matrix-assisted laser desorption ionization
time-of-flight MS.
| Spot | Protein name | Accession no. | Z-score score |
|---|
| 1 | Serotransferrin | P02787 | 61 |
| 2 | Heat shock | P01871 | 137 |
| 3 | Tubulin alpha
chain-like 3 | P02790 | 90 |
| 4 | Tropomyosin β
chain) | P01871 | 87 |
| 5 | Apolipoprotein E | P02768 | 136 |
Assessment of the reproducibility of this
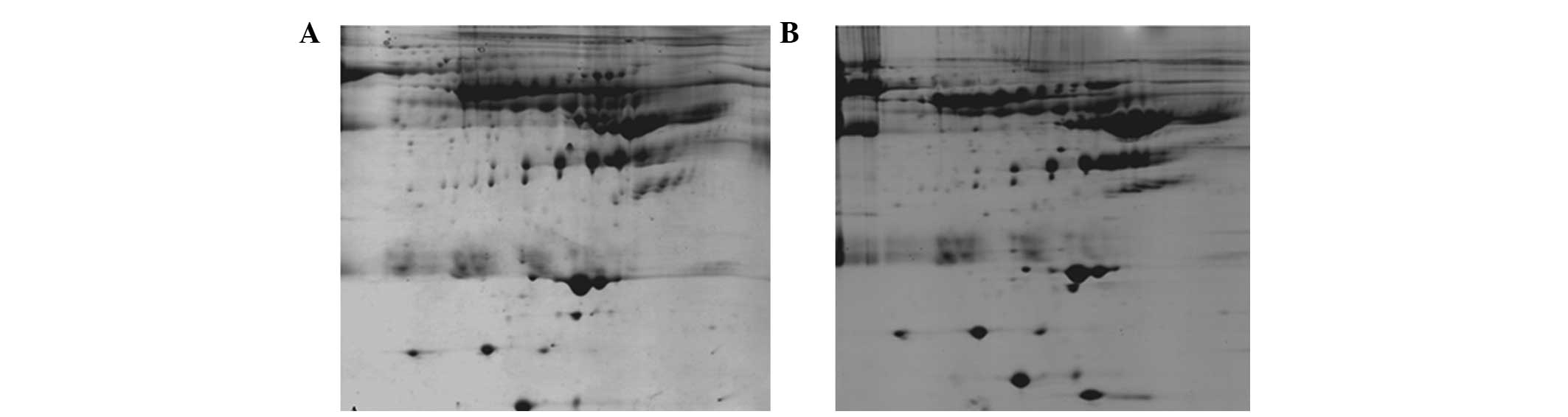
approach
To assess the repeatability of our approach, the
serum of two patients with colon cancer (age, 40–64; male:female,
1:1) was collected and the optimal protocol was applied. As shown
in Fig. 6, the 2D gels exhibited
low-abundance proteins and a reduction in abundance, thus
demonstrating that the approach had high repeatability.
Discussion
Given the importance of serum proteomics, this field
has progressed rapidly. Albumin and IgG represent ~80% of the total
protein in serum, thus the removal of albumin and IgG is expected
to increase the low-abundance protein load by ~4–5-fold (9). Therefore, removal of albumin and IgG
is adequate for the detection of primarily low abundance biomarkers
(10). Thus, the depletion of
highly abundant proteins is the main challenge of serum proteomics.
In addition, the solution obtained from high-abundance protein
deletion kits contains a rich array of compounds, which interfere
with IEF.
To date, numerous high-abundance protein depletion
kits have been designed for the removal of highly abundant proteins
from serum (11). Therefore,
selecting the most appropriate depletion kit for serum proteomics
is important. The selection depends on the balance between the
removal of highly abundant proteins and the loss of associated
low-abundance proteins. Affinity-based depletion kits are cheaper,
have a higher loading capacity and avoid cross-contamination. In
addition, this type of kit does not require any specific equipment,
thus it is used frequently.
Based on the analysis of all the methods, the
Cibacron Blue columns were selected for further investigation. The
ProteoPrep Blue Albumin and IgG Depletion kit and the Albumin and
IgG Removal kit are based on dye affinity and are commonly used. In
the present study, these two kits were compared with regards to
depletion efficiency, specificity and capacity. In addition, the
protocol for downstream IEF was optimized, which improves the
quality of the 2D gel. Finally, test reproducibility of this
approach was provided using the serum of different patients.
SDS PAGE and 2D gel clearly demonstrated that the
gel which applied the ProteoPrep Blue Albumin and IgG Depletion kit
had a higher quality, more protein spots and less albumin. This
demonstrated that the ProteoPrep Blue Albumin and IgG Depletion kit
could effectively remove high-abundance proteins.
The eluate following albumin and IgG depletion
demonstrated that this kit specifically depleted highly abundant
proteins. Identification of five unique proteins demonstrated that
they were all low-abundance proteins. These results further
demonstrated that this kit can specifically deplete high-abundance
proteins.
According to a previous study, the protein
concentration of the loading sample for silver stained 2-DE gels
should not be <0.5 µg/µl (10). Thus, a large quantity of protein
used in proteomics studies is often required (12). The present study demonstrated that
the ProteoPrep Blue Albumin and IgG Depletion kit can load a higher
volume and produce a higher protein yield, which meets the
requirement for downstream 2-DE.
In addition, the optimal approach to obtain the most
protein is through the simplest procedure. However, the majority of
commercial columns require additional steps to remove the
interferential materials (13)
following depletion of highly abundant proteins, as they use
alkaline solution or high level salt solution to elute the unbound
protein. They would interfere with IEF. The ProteoPrep Blue Albumin
and IgG Depletion kit has notable advantages. For example, the
solution obtained following depletion can be used for IEF directly
since the wash solution contains fewer ions.
In conclusion, of the two kits, the ProteoPrep Blue
Albumin and IgG Depletion kit was selected for two reasons.
Firstly, this kit had high depletion efficiency, specificity and
capacity. Secondly, the solution following depletion could be used
for IEF directly.
Normally, during IEF, the electric current must be
<50 µA, a higher electric current would obstruct IEF.
Thus, salt concentration is a critical factor of IEF as a high salt
concentration results in a high electric current. Prolonging the
low voltage process can remove salts. The present study initially
followed standard protocols of IEF [all low voltage steps running
for 1 (protocol A) or 2 h (protocol A and B)]. During IEF, the
electric current reached the limitation quickly. 2D gel produced
from protocol A and B with few protein spots also demonstrated that
these two protocols resulted in unsuccessful IEF. Therefore, low
voltage steps were prolonged to 3 h for protocol C, D and E. With
this modification, the current decreased to 30 µA when the
voltage reached 100 kvh, indicating that this modification is
useful for salt depletion. In addition, three types of final
voltage were compared: 100 k vhs (protocol C), 110 k vhs (protocol
D) and 130 k vhs (protocol E). 2D gel from protocol C and D
exhibited horizontal streaking and fewer protein spots, whereas the
2D gel of protocol C resulted in >500 protein spots. Overall,
protocol C obtains high quality 2D gel with less horizontal
streaking and more protein spots, and thus method C was selected
for further investigation.
For high reproducibility, an ideal research approach
is required. To confirm the reproducibility of our method, serum
was collected from two patients with colon cancer. These two
patients received radiotherapy and chemotherapy for >6 months.
Due to the side effects of the treatments, their serum protein is
unique, thus their serum was selected to assess the
reproducibility.
Albumin and IgG were depleted using the ProteoPrep
Blue Albumin and IgG Depletion kit. Subsequently, the solution
obtained underwent IEF with protocol C. Following this approach
using serum of patients with colon cancer, high quality 2D gels
were obtained, which have low-abundance proteins and reduced
high-abundance protein, demonstrating that this approach has high
reproducibility.
In conclusion, an effective approach for serum
proteomics was successfully provided. This included using the
ProteoPrep Blue Albumin and IgG Depletion kit to deplete albumin
and IgG, combined with protocol C to run IEF, which obtains high
quality 2D gels. In addition, five unique proteins were
successfully identified by MALDI-TOF/TOF MS and it was demonstrated
that they were all low-abundance proteins. The above results
demonstrated that our approach was highly effective with a high
reproducibility, subsequently paving the way for serum proteomics
research.
Acknowledgments
This study was supported by the National Science
Foundation of China (grant no. 81053424), the Natural Science
Foundation of Guangdong province (grant no. 2014A030313802), the
Bureau of Health of Guangzhou Municipality (grant nos. 20141A011016
and 20152A011010), and the Traditional Chinese Medicine Bureau of
Guangdong Province (grant no. 2014021).
References
|
1
|
Ahmed N, Barker G, Oliva K, Garfin D,
Talmadge K, Georgiou H, Quinn M and Rice G: An approach to remove
albumin for the proteomic analysis of low abundance biomarkers in
human serum. Proteomics. 3:1980–1987. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Anderson NL and Anderson NG: The human
plasma proteome: History, character, and diagnostic prospects. Mol
Cell Proteomics. 1:845–867. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bjorhall K, Miliotis T and Davidsson P:
Comparison of different depletion strategies for improved
resolution in proteomic analysis of human serum samples.
Proteomics. 5:307–317. 2005. View Article : Google Scholar
|
|
4
|
Desrosiers RR, Beaulieu E, Buchanan M and
Béliveau R: Proteomic analysis of human plasma proteins by
two-dimensional gel electrophoresis and by antibody arrays
following depletion of high-abundance proteins. Cell Biochem
Biophys. 49:182–195. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Echan LA, Tang HY, Ali-Khan N, Lee K and
Speicher DW: Depletion of multiple high-abundance proteins improves
protein profiling capacities of human serum and plasma. Proteomics.
5:3292–3303. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Greenough C, Jenkins RE, Kitteringham NR,
Pirmohamed M, Park BK and Pennington SR: A method for the rapid
depletion of albumin and immunoglobulin from human plasma.
Proteomics. 4:3107–3111. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hoffman SA, Joo WA, Echan LA and Speicher
DW: Higher dimensional (Hi-D) separation strategies dramatically
improve the potential for cancer biomarker detection in serum and
plasma. J Chromatogr B Analyt Technol Biomed Life Sci. 849:43–52.
2007. View Article : Google Scholar
|
|
8
|
Kim MR and Kim CW: Human blood plasma
preparation for two-dimensional gel electrophoresis. J Chromatogr B
Analyt Technol Biomed Life Sci. 849:203–210. 2007. View Article : Google Scholar
|
|
9
|
Lescuyer P, Hochstrasser D and Rabilloud
T: How shall we use the proteomics toolbox for biomarker discovery?
J Proteome Res. 6:3371–3376. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Liu B, Qiu FH, Voss C, Xu Y, Zhao MZ, Wu
YX, Nie J and Wang ZL: Evaluation of three high abundance protein
depletion kits for umbilical cord serum proteomics. Proteome Sci.
9:242011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Mahn A, Reyes A, Zamorano M, Cifuentes W
and Ismail M: Depletion of highly abundant proteins in blood plasma
by hydrophobic interaction chromatography for proteomic analysis. J
Chromatogr B Analyt Technol Biomed Life Sci. 878:1038–1044. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Qiu F, Huang D, Xiao H, Qiu F, Lu L and
Nie J: Detection of tyrosine phosphorylated proteins in
hepatocellular carcinoma tissues using a combination of
GST-Nck1-SH2 pull-down and two-dimensional electrophoresis. Mol Med
Rep. 7:1209–1214. 2013.PubMed/NCBI
|
|
13
|
Thadikkaran L, Siegenthaler MA, Crettaz D,
Queloz PA, Schneider P and Tissot JD: Recent advances in
blood-related proteomics. Proteomics. 5:3019–3034. 2005. View Article : Google Scholar : PubMed/NCBI
|