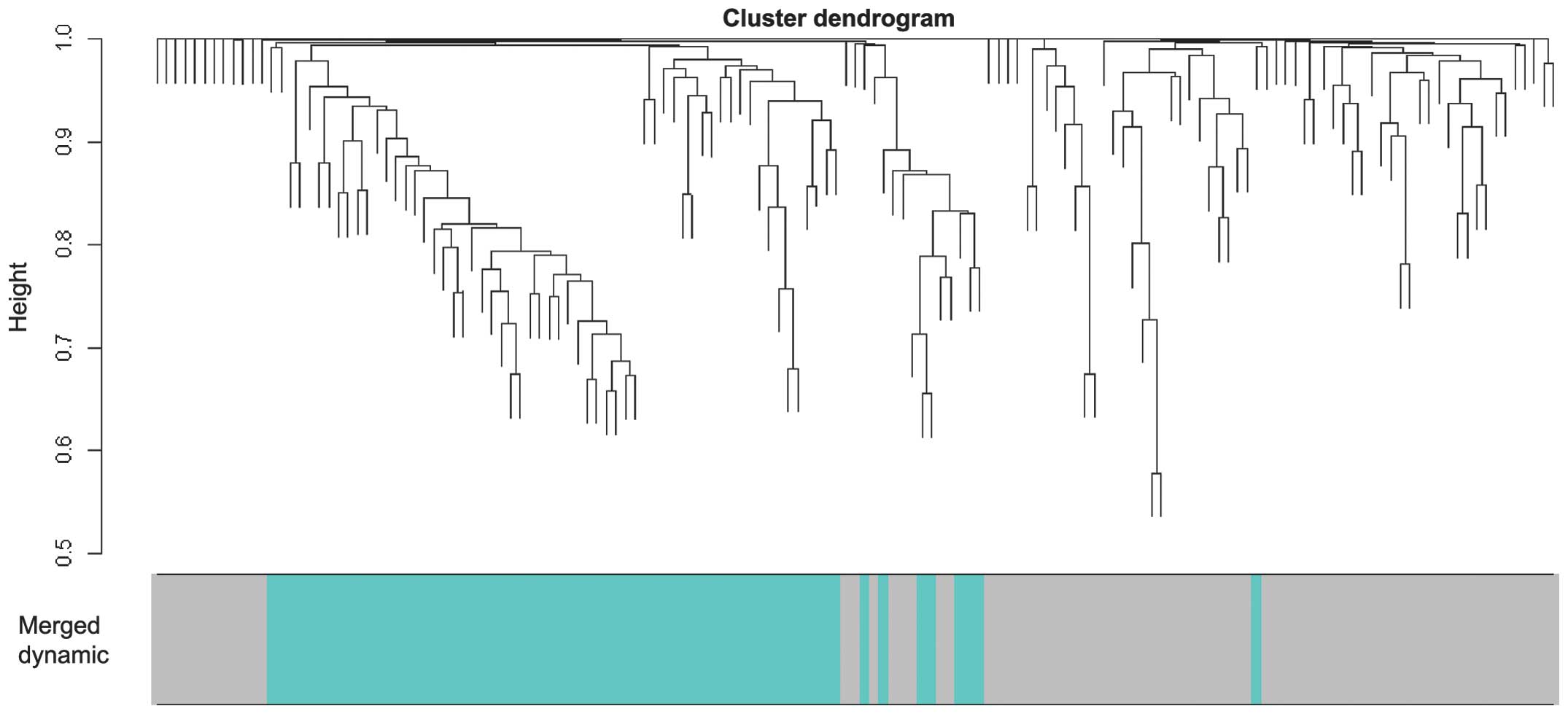
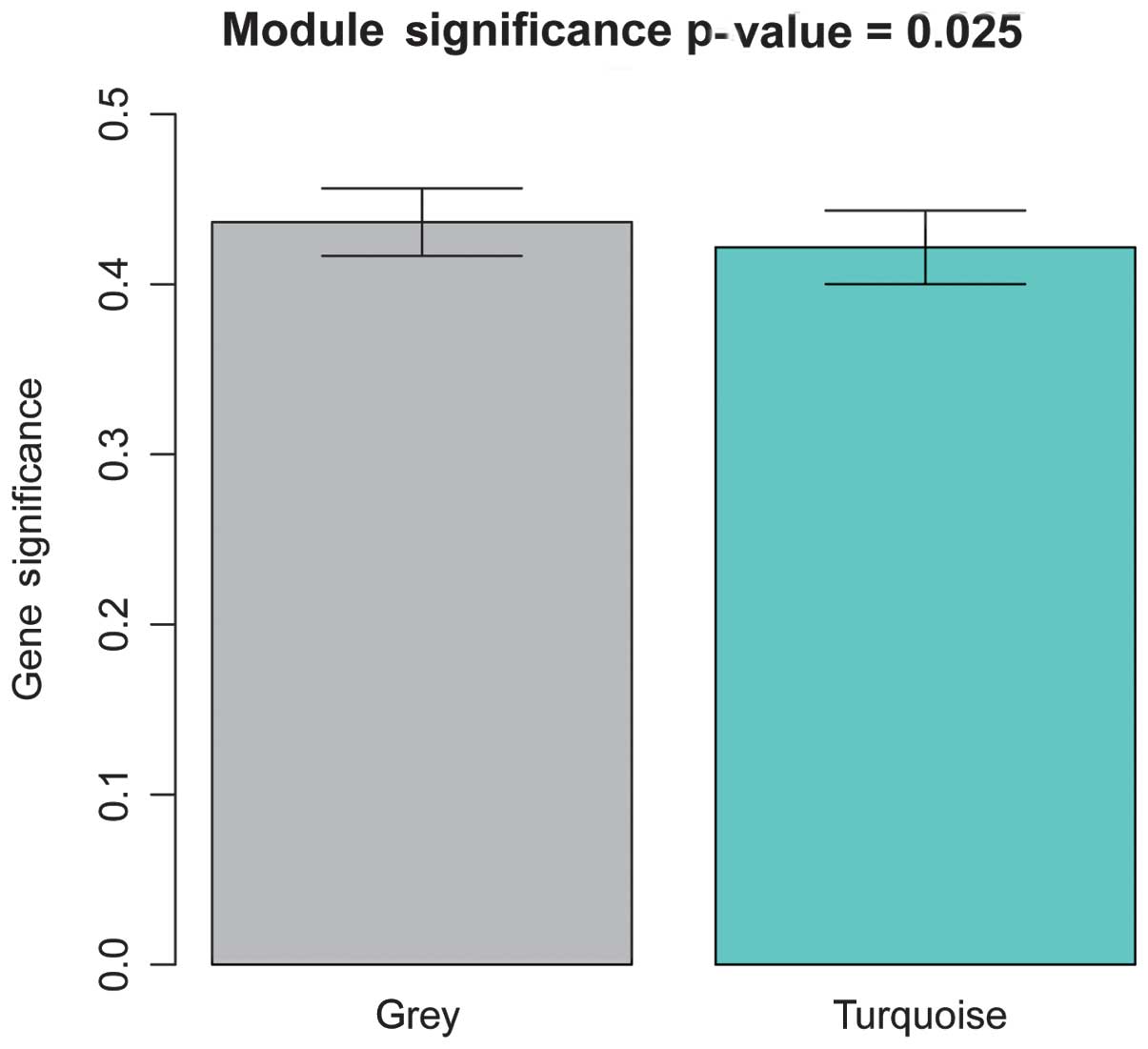
|
1
|
Hiatt WR and Nehler MR: Peripheral
arterial disease. 2007
|
|
2
|
Hiatt WR: Medical treatment of peripheral
arterial disease and claudication. N Engl J Med. 344:1608–1621.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cronberg C, Sjöberg S, Albrechtsson U,
Leander P, Lindh M, Norgren L, Danielsson P, Sonesson B and Larsson
EM: Peripheral arterial disease. Contrast-enhanced 3D MR
angiography of the lower leg and foot compared with conventional
angiography. Acta Radiol. 44:59–66. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ouriel K: Peripheral arterial disease.
Lancet. 358:1257–1264. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wilson AM, Kimura E, Harada RK, Nair N,
Narasimhan B, Meng XY, Zhang F, Beck KR, Olin JW, Fung ET and Cooke
JP: Beta2-Microglobulin as a biomarker in peripheral arterial
disease: Proteomic profiling and clinical studies. Circulation.
116:1396–1403. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Smadja DM, D'audigier C, Bièche I, Evrard
S, Mauge L, Dias JV, Labreuche J, Laurendeau I, Marsac B, Dizier B,
et al: Thrombospondin-1 is a plasmatic marker of peripheral
arterial disease that modulates endothelial progenitor cell
angiogenic properties. Arterioscler Thromb Vasc Biol. 31:551–559.
2011. View Article : Google Scholar
|
|
7
|
Busti C, Falcinelli E, Momi S and Gresele
P: Matrix metalloproteinases and peripheral arterial disease.
Intern Emerg Med. 5:13–25. 2010. View Article : Google Scholar
|
|
8
|
Masud R, Shameer K, Dhar A, Ding K and
Kullo IJ: Gene expression profiling of peripheral blood mononuclear
cells in the setting of peripheral arterial disease. J Clin
Bioinforma. 2:62012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Horvath S and Dong J: Geometric
interpretation of gene coexpression network analysis. PLoS Comput
Biol. 4:e10001172008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ruan J, Dean AK and Zhang W: A general
co-expression network-based approach to gene expression analysis:
Comparison and applications. BMC Syst Biol. 4:82010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Malki K, Tosto MG, Jumabhoy I, Lourdusamy
A, Sluyter F, Craig I, Uher R, McGuffin P and Schalkwyk LC:
Integrative mouse and human mRNA studies using WGCNA nominates
novel candidate genes involved in the pathogenesis of major
depressive disorder. Pharmacogenomics. 14:1979–1990. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Udyavar AR, Hoeksema MD, Clark JE, Zou Y,
Tang Z, Li Z, Li M, Chen H, Statnikov A, Shyr Y, et al:
Co-expression network analysis identifies spleen tyrosine kinase
(SYK) as a candidate oncogenic driver in a subset of small-cell
lung cancer. BMC Syst Biol. 7(Suppl 5): S12013. View Article : Google Scholar
|
|
13
|
Zhao H, Cai W, Su S, Zhi D, Lu J and Liu
S: Screening genes crucial for pediatric pilocytic astrocytoma
using weighted gene coexpression network analysis combined with
methylation data analysis. Cancer Gene Ther. 21:448–455. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Barrett T, Wilhite SE, Ledoux P,
Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH,
Sherman PM, Holko M, et al: NCBI GEO: Archive for functional
genomics data sets-update. Nucleic Acids Res. 41:D991–D995,
Database issue. 2013. View Article : Google Scholar
|
|
15
|
Gautier L, Cope L, Bolstad BM and Irizarry
RA: affy-analysis of affymetrix genechip data at the probe level.
Bioinformatics. 20:307–315. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Smyth GK: Limma: Linear models for
microarray data. Bioinformatics and Computational Biology Solutions
Using R and Bioconductor. Gentleman R, Carey V, Huber W, Irizarry R
and Dudoit S: Springer; New York: pp. 397–420. 2005, View Article : Google Scholar
|
|
17
|
Yip AM and Horvath S: Gene network
interconnectedness and the generalized topological overlap measure.
BMC Bioinformatics. 8:222007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Langfelder P, Zhang B and Horvath S:
Defining clusters from a hierarchical cluster tree: The dynamic
tree cut package for R. Bioinformatics. 24:719–720. 2008.
View Article : Google Scholar
|
|
19
|
Langfelder P and Horvath S: WGCNA: An R
package for weighted correlation network analysis. BMC
Bioinformatics. 9:559–571. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Huang Da W, Sherman BT, Tan Q, Collins JR,
Alvord WG, Roayaei J, Stephens R, Baseler MW, Lane HC and Lempicki
RA: The DAVID gene functional classification tool: A novel
biological module-centric algorithm to functionally analyze large
gene lists. Genome Biol. 8:R1832007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wu J, Mao X, Cai T, Luo J and Wei L: KOBAS
server: A web-based platform for automated annotation and pathway
identification. Nucleic Acids Res. 34:W720–W724. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
American Disease Association: Peripheral
arterial disease in people with diabetes. J Am Podiatr Med Assoc.
95:309–319. 2005. View
Article : Google Scholar
|
|
24
|
Gartel AL and Radhakrishnan SK: Lost in
transcription: p21 repression, mechanisms and consequences. Cancer
Res. 65:3980–3985. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Cazzalini O, Scovassi AI, Savio M, Stivala
LA and Prosperi E: Multiple roles of the cell cycle inhibitor
p21(CDKN1A) in the DNA damage response. Mutat Res. 704:12–20. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Gulbis JM, Kelman Z, Hurwitz J, O'donnell
M and Kuriyan J: Structure of the C-terminal region of
p21(WAF1/CIP1) complexed with human PCNA. Cell. 87:297–306. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Rodriguez I, Coto E, Reguero JR, González
P, Andrés V, Lozano I, Martín M, Alvarez V and Morís C: Role of the
CDKN1A/p21, CDKN1C/p57 and CDKN2A/p16 genes in the risk of
atherosclerosis and myocardial infarction. Cell Cycle. 6:620–625.
2007. View Article : Google Scholar
|
|
28
|
Abate C, Luk D, Gagne E, Roeder RG and
Curran T: Fos and jun cooperate in transcriptional regulation via
heterologous activation domains. Mol Cell Biol. 10:5532–5535. 1990.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hunter T: Oncoprotein networks. Cell.
88:333–346. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Braun-Dullaeus RC, Mann MJ and Dzau VJ:
Cell cycle progression New therapeutic target for vascular
proliferative disease. Circulation. 98:82–89. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Mahner S, Baasch C, Schwarz J, Hein S,
Wölber L, Jänicke F and Milde-Langosch K: C-Fos expression is a
molecular predictor of progression and survival in epithelial
ovarian carcinoma. Br J Cancer. 99:1269–1275. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Bossis G, Malnou CE, Farras R,
Andermarcher E, Hipskind R, Rodriguez M, Schmidt D, Muller S,
Jariel-Encontre I and Piechaczyk M: Down-regulation of c-Fos/c-Jun
AP-1 dimer activity by sumoylation. Mol Cell Biol. 25:6964–6979.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Chenevard R, Hürlimann D, Béchir M,
Enseleit F, Spieker L, Hermann M, Riesen W, Gay S, Gay RE, Neidhart
M, et al: Selective COX-2 inhibition improves endothelial function
in coronary artery disease. Circulation. 107:405–409. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Liou JY, Deng WG, Gilroy DW, Shyue SK and
Wu KK: Colocalization and interaction of cyclooxygenase-2 with
caveolin-1 in human fibroblasts. J Biol Chem. 276:34975–34982.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhu H, Yue J, Pan Z, Wu H, Cheng Y, Lu H,
Ren X, Yao M, Shen Z and Yang JM: Involvement of Caveolin-1 in
repair of DNA damage through both homologous recombination and
non-homologous end joining. PloS One. 5:e120552010. View Article : Google Scholar : PubMed/NCBI
|