Introduction
Epidemiological and case control studies have
consistently indicated that moderate and mild elevation of plasma
homocysteine (Hcy), an intermediate metabolite of methionine, is an
important and independent risk factor in the development of
atherosclerosis (AS) (1–3). Hcy can induce endothelial
dysfunction, foam formation and promote vascular smooth muscle cell
(VSMC) proliferation (4–6). It is known that VSMC proliferation is
pivotal in the pathogenesis and progression of AS, and numerous
studies have been performed to elucidate the detailed mechanisms
and relevant genes, including platelet-derived growth factor,
phosphatase and tensin homolog and p53, involved in the
proliferation of VSMCs (7–9). However, the exact mechanisms that are
responsible for Hcy-induced VSMC proliferation remain to be fully
elucidated.
MicroRNAs (miRNAs) are short (19–25 nucleotide),
non-coding RNAs, which regulate the expression of genes at the
post-transcriptional level by binding to the 3′ untranslated region
(UTR) of target mRNAs, leading to mRNA degradation or translation
repression (10). Previous
evidence has demonstrated that miRNAs are key in the progression of
AS, the involvement of which is predominantly associated with
controlling the basic function of endothelial cells, VSMCs and
macrophages (11). Its expression
is modulated by different stimuli involved in every stage of AS.
Notably, a group of miRNAs, including miR-143/145, miR-221/222,
miR-24, miR-146a and miR-21, have been identified to modulate VSMC
differentiation, phenotypic switch and neointimal formation
following vascular injury (12,13).
Among these, miR-143 is a markedly enriched miRNA in VSMCs, which
has been investigated extensively in the field of VSMC biology. The
expression of miR-143 is critical to the modulation of VSMC
phenotype, by promoting differentiation and repressing the
proliferation of VSMCs (14).
Considerable evidence has shown that the dysregulation of miRNA
expression involved in cancer can be triggered by multiple
mechanisms, including aberrant DNA methylation of the miRNA
promoter (15). However, the
underlying mechanisms of miRNA dysregulation in VSMC proliferation
resulting from hyperhomocysteinemia remain to be elucidated.
DNA methylation is a major epigenetic factor
regulating genome reprogramming, cell differentiation and
developmental gene expression, which is catalyzed by a family of
DNA methyltransferases (DNMTs), including DNMT1, DNMT3a and DNMT3b
(16,17). DNMT1 contributes to the maintenance
of DNA methylation patterns, whereas DNMT3a/3b contribute to de
novo methylation (18). Hcy is
a sulfhydryl-containing amino acid, which is either derived from
the metabolic demethylation of methionine or remethylated to
methionine (19). Additionally,
the hypermethylation of gene promoters is a common mechanism of
miRNA silencing at the transcriptional level (20). Previous studies have shown that
miRNAs can be involved in the promoter methylation of CpG islands
by targeting the DNMT 3′UTR (21,22).
In addition, our previous study found that DNMT3a is one potential
target of miR-143 using bioinformatics analysis. Therefore, whether
miR-143 methylation regulated by DNMT3a mediates Hcy-induced VSMC
proliferation requires clarification.
The present study aimed to identify whether DNA
methylation was involved in the downregulation of miR-143
expression and whether there was a circular regulation loop between
miR-143 and DNMT3a. The results provided novel insights into the
molecular mechanism underlying Hcy-induced VSMC proliferation.
Materials and methods
Cell culture
Primary cultured VSMCs were obtained from human
umbilical vein media (Department of Obstetrics, General Hospital of
Ningxia Medical University (Yinchuan, China). The present study was
approved by the ethics committee of the Ningxia Medical University.
The VSMCs were maintained in Dulbecco's modified Eagle's
Medium-Han's F12media (GE Healthcare Life Sciences, Logan, UT, USA)
supplemented with 20% fetal calf serum (FCS; Gibco; Thermo Fisher
Scientific, Inc., Waltham, MA, USA), 100 U/ml penicillin and 100
U/ml streptomycin (Sigma-Aldrich, St. Louis, MO, USA) at 37°C in an
incubator with 5% CO2. The VSMCs were grow for 3–5
generations and seeded into 6-well plates at a density of
1×106 cells/well. Hcy (Sigma-Aldrich) was added to the
cells at concentrations of 0 (control), 50, 100, 200 and 500
µM, and 100 µM Hcy+30 µM folate
(Sigma-Aldrich), respectively, in order to compensate for the short
half-life of homocysteine at 37°C in an incubator containing 5%
CO2, which was replenished every 8 h, three times in
total.
Cell proliferation assay
Cell proliferation was measured using a
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)
assay (Nanjing Jiancheng Bioengineering Institute, Nanjing, China).
The VSMCs were seeded in 96-well plates at a density of
103–104 cells/well, and incubated with 0, 50,
100, 200, 500 µM concentrations of Hcy and 100 µM
Hcy+30 µM folate for 72 h. Subsequently, 20 µl MTT (5
mg/ml) was added to each well and incubated at 37°C for 4 h. The
medium was removed, and 150 µl dimethyl sulfoxide
(Sigma-Aldrich) was added to each well to resuspend the MTT
metabolic product. Following 10 min of incubation at room
temperature, the absorbance of the dissolved formazan was measured
at 490 nm (A490) using a scanning microplate spectrophotometer
(BioTek Epoch; BioTek Instruments, Inc., Winooski, VT, USA).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) detection of the mRNA
expression levels of miR-143 and DNMT3a in VSMCs
Total RNA, containing small RNA, was extracted from
the VSMCs using TRIzol reagent (Invitrogen; Thermo Fisher
Scientific, Inc.), according to the manufacturer's protocol. For
miRNA qPCR, prior RT was performed using a Revertid™ First Strand
cDNA Synthesis kit (Thermo Fisher Scientific, Inc.). For RT, 1
µg of RNA containing miRNA was polyadenylated by poly (A)
polymerase and then reverse transcribed to cDNA using oligo-dT
primers. The cDNA (2 µl) then served as the template and was
added to 1 µl primers for SYBR real-time PCR using
TransStart® Top Green qPCR SuperMix (Transgen Biotech
Co, Ltd., Beijing, China). The sequence of the miR-143 specific
forward primer (Sangon Biotech Co., Ltd., Shanghai, China) was
5′-GGGTGAGATGAAGCACTGTAGCTC-3′ and the reverse primer was
5′-GCTGTCAACATACGCTACGTAACG-3′. Human U6 was used for
normalization. For DNMT3a mRNA qPCR, cDNA was synthesized with
oligo-dT primers and MMLV reverse transcriptase (Promega
Corporation, Madison, WI, USA), according to the manufacturer's
protocol. GAPDH was used as an endogenous control, and the primer
sequences (Sangon Biotech Co., Ltd.) were as follows: DNMT3a,
forward 5′-GGGGACGTCCGCAGCGTCACAC-3′ and reverse
5′-CAGGGTTGGACTCGAGAAATCGC-3′; and GAPDH, forward
5′-AGAAGGCTGGGGCTCATTTG-3 and reverse 5′-AGGGGCCATCCACAGTCTTC-3′.
All qPCR assays were performed using an FTC3000 real-time PCR
detection system (FengLing, Shanghai, China) with the following
program: 45 cycles at 95°C for 45 sec, 58°C for 45 sec and
extension 60°C for 60 sec. The relative quantification of the PCR
products was performed according to the 2−ΔΔcq method
(23) and normalized by the
control.
Western blot analysis
The cultured VSMCs were harvested using a protein
extraction kit (Nanjing KeyGen Biotech Co., Ltd., Nanjing, China).
A Bicinchoninic Acid kit (Sigma-Aldrich) was used to determine
protein concentration. Equal quantities of proteins (30 µg)
were separated by 10% SDS-PAGE and then transferred onto a
polyvinylidene fluoride membrane (EMD Millipore, Billerica, MA,
USA) following which the membrane was blocked in 5% nonfat milk for
2 h at room temperature. The membrane was washed following blocking
and then incubated overnight at 4°C with rabbit polyclonal
anti-DNMT3a antibody (1:5,000; Abcam, Cambridge, MA, USA; cat. no.
ab23565), and washed three time with phosphate-buffered saline with
0.5% Tween 20 (PBST; pH 4.0) for 5 min at room temperature. The
membranes were then incubated for 2 h at room temperature with goat
anti-rabbit horseradish peroxidase-conjugated IgG secondary
antibody (1:1,000; Novoprotein, Shanghai, China; cat. no. AB501).
Following washing again three times with PBST, the immunoreactive
protein bands were detected using enhanced chemiluminescence
solution (Beyotime Institute of biotechnology, China), and the
relative protein levels were determined by band densitometry using
a gel imaging system (Bio-Rad Laboratories, Inc., Hercules, CA,
USA).
Nested methylation-specific (nMS)-PCR for
determination of miR-143 methylation in VSMCs
A Wizard® Genomic DNA Purification kit
(Promega Corporation) was used to extract Genomic DNA from the
VSMCs. An EZ DNA Methylation-Gold kit (Zymo Research, Irvine, CA,
USA) was used to integrate DNA denaturation and bisulfite
conversion processes. The nMS-PCR consisted of a two-step PCR
amplification following standard sodium bisulfite DNA modification.
The first step of nMS-PCR was performed using the following outer
primer pair: Forward 5′-AGGAGTTTTAGATTAGTTTGGGTAA-3′ and reverse
5′-TTTTAAAACAAAATCTCCCTCTATC-3′. The second step of the nMS-PCR was
performed using the following PCR primers: Methylation primer,
forward 5′-ATTAGTTAGGTATGGTGGTGTACGT-3′ and reverse
5′-TTTAAAACAAAATCTCCCTCTATCG-3′; and unmethylation primer, forward
5′-ATTAGTTAGGTATGGTGGTGTATGT-3′ and reverse
5′-TTAAAACAAAATCTCCCTCTATCAC-3′. To reduce mispriming and to
increase efficiency, nMS-PCR was used for amplification. A total of
4 µl DNA, 12.5 µl buffer, and 1 µl primers
were subjected to 30 cycles in an nMS-PCR program (94°C for 30 sec;
66°C for 30 sec and 72°C for 1 min), followed by a 0.5°C decrease
in the annealing temperature every cycle. Following completion of
the nMS-PCR program, 20 cycles were subsequently run (94°C for 45
sec, 51°C for 45 sec and 72°C for 45 sec). Following amplification,
2% agarose gels containing ethidium bromide (Invitrogen; Thermo
Fisher Scientific, Inc.) were used for electrophoresis to separate
the PCR products. DNA bands were visualized by ultraviolet light
using the Bio-Rad gel imagine system, and the percentage of
methylation was calculated using the following formula: Methylation
(%) = methylation / (methylation + unmethylation) × 100%.
Cell transfection
The VSMCs were seeded into 6-well plates at a
density of 1×106 cells/well 1 day prior to transfection.
The cells were transiently transfected with pre-miR-143 and an
miR-143 inhibitor (Genepharma Co., Ltd., Shanghai, China),
DNMT3a-specific small interfering (si)RNA (Genepharma Co., Ltd.)
using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific,
Inc.) according to the manufacturer's protocol. Control-siRNA
(Genepharma Co., Ltd.) was co-transfected as a negative control at
37°C in an incubator containing 5% CO2. After 48 h, the
cells were collected for total RNA and protein extraction.
Construction of the luciferase reporter
vector targeting the DNMT3a 3′-UTR
For the luciferase reporter experiments, a DNMT3a
3′-UTR (Genepharma Co., Ltd.) segment of 611 bp, which contains the
potential binding site of miR-143, was amplified using PCR from 2
µl human genomic DNA and 1 µl primers, and inserted
into the pGL3-control vector with the simian virus 40 promoter and
enhancer (Promega Corporation) by using the XbaI site
immediately downstream from the stop codon of luciferase. The
following sets of primers were used to generate the specific
fragments: DNMT3a-UTR, forward 5′-GCTCTAGAACTGGCTACTGCTCTGTG-3′;
and reverse 5′-GCTCTAGAGAAATGTATGTCTGTCCCT-3′. The underlined
sequences indicate the endonuclease restriction site. The cycling
conditions included initial denaturation at 95°C for 5 min,
followed by 40 cycles of 95°C for 10 sec, 56°C for 30 sec and 72°C
for 30 sec, and then extension at 72°C for 10 min. When the
amplified 3′-UTR of DNMT3a, containing the predicted match seed of
miR-143, was cloned into the pGL3 vectors, the constructed plasmid,
pGL3-DNMT3a, was used to perform the subsequent luciferase
assay.
Statistical analysis
GraphPad Prism 5.0 software (GraphPad Software,
Inc., La Jolla, CA, USA) was used for statistical analysis. The
results are expressed as the mean ± standard deviation from at
least three independent experiments. The data were analyzed using
one-way analysis of variance, and were further analyzed using the
Student Newman-Keuls test for multiple comparisons within treatment
groups or paired Student's t-test for two groups. P<0.05 was
considered to indicate a statistically significant difference.
Results
Expression of miR-143 and its effect on
the proliferation of VSMCs induced by Hcy
The effect of Hcy on VSMC proliferation was detected
using an MTT assay. As shown in Fig.
1A, VSMC proliferation increased by 1.39-fold in the 100
mM Hcy group, compared with the control group (P<0.05).
By contrast, VSMC proliferation decreased almost 27% in the 100
µM Hcy+folate group, compared with the 100 µM Hcy
group (P<0.01). These results confirmed that Hcy stimulated VSMC
proliferation, as reported previously (24), whereas folate exerted a marked
antagonistic effect. In addition, the present study detected the
expression of miR-143 induced by Hcy in the VSMCs. As shown in
Fig. 1B, the levels of miR-143
were significantly downregulated in the Hcy group, particularly in
the 100 µM Hcy group, compared with the control group
(P<0.01). By contrast, the expression of miR-143 was markedly
upregulated in response to 100 µM Hcy+folate, compared with
the 100 µM group (P<0.01). To further confirm the role of
miR-143 in the proliferation of VSMCs induced by Hcy, pre-miR-143
and miR-143 inhibitor were transfected into VSMCs, respectively, to
measure cell proliferation. As shown in Fig. 1C, compared with control group, the
expression levels of miR-143 were significantly increased in the
pre-miR-143 group (P<0.01), and significantly decreased in the
miR-143 inhibitor group (P<0.01), compared with the control. The
proliferation of the VSMCs was increased significantly in the
pre-miR-143 transfected cells, whereas the miR-143 inhibitor
reduced the levels of VSMC proliferation, compared with the
control. In addition, the proliferation of the VSMCs showed a
significant increased in the VSMCs treated with 100 µM Hcy
(Fig. 1D). These results
demonstrated that the downregulation of miR-143 promoted the
proliferation of VSMCs induced by Hcy.
 | Figure 1Expression of miR-143 and its effect
on the proliferation in VSMCs induced by Hcy. (A) Effect of Hcy on
VSMC proliferation, evaluated using an MTT assay. The VSMCs were
incubated with 0, 50, 100, 200, 500 µM Hcy and 100 µM
Hcy+30 µM folate for 72 h. (B) Relative levels of miR-143 in
VSMCs induced by Hcy were analyzed using reverse
transcription-quantitative polymerase chain reaction. Expression of
miR-143 was normalized to U6. (C) Relative levels of miR-143 in
VSMCs transfected with pre-miR-143 and miR-143 inhibitor. (D)
Proliferation of VSMCs transfected with pre-miR-143 and miR-143
inhibitor after 48 h were measured using an MTT assay. UV-visible
absorbance was measured at 490 nm. Data are expressed as the mean ±
standard deviation (n=3). *P<0.05 and
**P<0.01, compared with the control group;
∆P<0.01, compared with the 100 µM Hcy group.
miR, microRNA; MTT,
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide; VSMC,
vascular smooth muscle cell; Hcy, homocysteine. |
Downregulation of miR-143 is caused by
DNA hypermethylation in VSMCs induced by Hcy
In order to evaluate the mechanism responsible for
the downregulation in the expression of miR-143 in the VSMC, the
present study measured the level of methylation in the DNA region
encoding miR-143 using nMS-PCR. As shown in Fig. 2, the methylation levels of miR-143
in the 100 and 200 µM Hcy groups were significantly
increased, by 4.49- and 2.64-fold, respectively, compared with the
control group, whereas the methylation levels of miR-143 decreased
by 48% in the 100 µM Hcy+folate group, compared with the 100
µM Hcy group (P<0.01). These results indicated that the
early downregulation in the production of miR-143 in the
Hcy-induced VSMCs was provoked by DNA hypermethylation in the
region encoding this microRNA.
 | Figure 2Methylation level of miR-143,
detected using nMS-PCR. miR-143 was amplified from
bisulfite-converted DNA and analyzed using nMS-PCR. Results
represent the mean of three quantifications performed in duplicate,
and are expressed as the percentage methylation. Data are expressed
as the mean ± standard deviation (n=3). *P<0.05 and
**P<0.01, compared with the control group;
∆P<0.01, compared with the 100 µM Hcy group.
M, amplified by methylation-specific primer; U, amplified by
unmethylation-specific primer; miR, microRNA; VSMC, vascular smooth
muscle cell; Hcy, homocysteine; nMS-PCR, nested
methylation-specific polymerase chain reaction. |
DNMT3a upregulation is responsible for
hypermethylation of miR-143 gene promoters in VSMCs induced by
Hcy
To investigate whether the hypermethylation of
miR-143 was associated with altered expression levels of DNMT3a in
VSMCs, the present study determined the expression levels of DNMT3a
using RT-qPCR and western blot analyses. As shown in Fig. 3A, the mRNA levels of DNMT3a in the
50, 100 and 200 µM Hcy groups were increased by 4.28-,
5.82-, 4.89-fold, respectively, compared with the control group
(P<0.05). By contrast, the mRNA levels of DNMT3a in the 100
µM Hcy+folate group decreased to 45% of that in the 100
µM Hcy group (P<0.01). In addition, compared with the
control group, the protein level of DNMT3a in the 100 µM Hcy
group was significantly increased, by 1.45-fold, however, in the
100 µM Hcy+folate group, the protein expression of DNMT3a
decreased to 41% of that in the 100 µM Hcy group (P<0.01;
Fig. 3B). To further verify the
effect of DNMT3a on miR-143 hypermethylation, the VSMCs were
transfected with a DNMT3a siRNA plasmid for 48 h. The mRNA and
protein levels of DNMT3a decreased significantly in the siDNMT3a
group, compared with the control or negative controls (P<0.01;
Fig. 3C and D). The results
suggested a significant reduction in the methylation level of
miR-143 following transfection of the VSMCs with DNMT3a siRNA
(P<0.01; Fig. 3E), which
demonstrated that DNMT3a was important in miR-143 hypermethylation
induced by Hcy.
 | Figure 3DNMT3a is involved in miR-143
hypermethylation. (A) mRNA levels of DNMT3a in VSMCs were detected
using reverse transcription-quantitative PCR, with GAPDH as an
internal control. (B) Protein levels of DNMT3a in VSMCs were
determined using western blot analysis, with β-actin as a loading
control. The (C) mRNA and (D) protein expression levels of DNMT3a
in VSMCs transfected with the DNMT3a siRNA plasmid for 48 h. (E)
miR-143 methylation levels in VSMCs following siDNMT3a transfection
were analyzed using nested methylation-specific PCR. Data are
expressed as the mean ± standard deviation (n=3).
*P<0.05 and **P<0.01, compared with the
control group; ∆P<0.01, compared with the 100
µM Hcy group. M, amplified by methylation-specific primer;
U, amplified by unmethylation-specific primer; DNMT, DNA
methyltransferase; miR, microRNA; si, small interfering; VSMC,
vascular smooth muscle cell; Hcy, homocysteine; PCR, polymerase
chain reaction. N-controls, negative controls. |
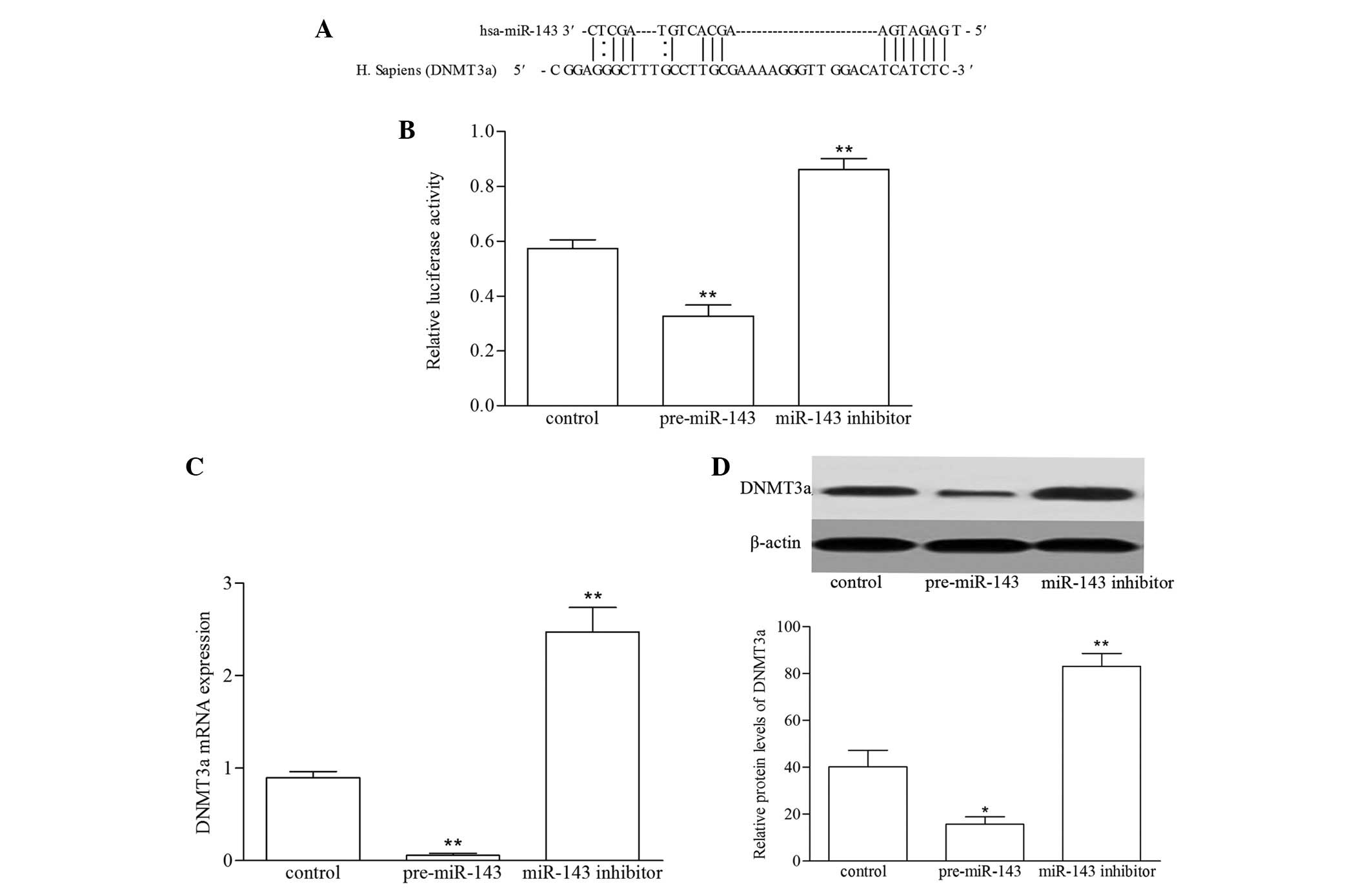
miR-143 targets DNMT3a in VSMCs
In order to identify the possible regulatory pathway
of miR-143 targeting DNMT3a in VSMCs, the sequence alignment of
human miR-143 with human DNMT3a 3′UTR was conserved (Fig. 4A) (25). The sequence was cloned into the
3′-UTR of the firefly luciferase gene and co-transfected with the
pre-miR-143 and miR-143 inhibitor, following which the luciferase
activities were assayed using a luciferase reporter system. As
shown in Fig. 4B, luciferase
activity was significantly reduced in the VSMCs transfected with
pre-miR-143, and was increased in the VSMCs transfected with
miR-143 inhibitor (P<0.01). To assess whether the expression of
DNMT3a was regulated by miR-143, RT-qPCR and western blot analyses
were performed to determine the mRNA and protein levels of DNMT3a
following transfection of the VSMCs with the pre-miR-143 and
miR-143 inhibitor. As shown in Fig. 4C
and D, enforced miR-143 expression led to a reduction in the
expression of DNMT3a at the mRNA and protein levels, compared with
the control group. By contrast, the inhibition of miR-143 in the
VSMCs increased the expression of DNMT3a. These results
demonstrated that miR-143 directly targeted DNMT3a by binding their
3′-UTRs, thus suppressing protein expression.
Discussion
Accumulating evidence has suggested that Hcy is an
independent risk factor of AS, although the underlying mechanism
remains to be fully elucidated (26). In particular, evidence has
indicated that the proliferation of VSMCs is important in the
development of AS (27). The
present study showed that Hcy had a stimulatory effect on VSMC
proliferation, which was consistent with these previous studies.
However, the underlying mechanism accounting for the promotion of
VSMC proliferation by Hcy remains to be fully elucidated.
In previous years, emerging evidence has
demonstrated that miRNAs are pivotal in the control of VSMC
proliferation and differentiation (28). Several studies have demonstrated
that miR-143 is involved in the phenotypic modulation of VSMCs and
cardiovascular diseases (29,30).
The results obtained from RT-qPCR validation in the present study
showed that miR-143 was downregulated in VSMCs induced by Hcy,
particularly at a concentration of 100 µM Hcy, which
exhibited the most marked proliferating effect on the VSMCs. In
addition, the VSMCs transfected pre-miR-143 and miR-143 inhibitor
further confirmed the suppressive effect of miR-143 on VSMC
proliferation, which had the same effect as folate. In the
'methionine cycle', Hcy can be converted to methionine by acquiring
a methyl group from N-5-methyltetrahydrofolate by methionine
synthase, and be subsequently activated to S-adenosyl methionine
(SAM) (31). Folate supply
increases the production of N-5-methyltetrahydrofolate, promoting
the transformation of Hcy to SAM (32). Therefore, the addition of folate in
the present study had an antagonistic effect on the deteriorative
roles of Hcy. In the present study, the expression of miR-143 was
downregulated in the Hcy group, which attenuated its suppressive
effect and promoted VSMC proliferation.
Increasing evidence has shown that DNA
hypermethylation is one cause of inactivation for numerous genes,
and has been identified more recently as a cause of the
inactivation of miRNA gene expression (33). Hcy is an intermediate product in
transmethylation reactions, including DNA methylation modification,
which is involved in the epigenetic regulation of gene expression
(34). Therefore, the present
study further evaluated the mechanism responsible for the
downregulated expression of miR-143 in the VSMCs. As the results of
the nMS-PCR analysis revealed, there was an increase in the level
of methylation in the miR-143-coding region, however, its
methylation level was decreased in the folate group, due to the
reverse effects of folate against Hcy. Combining the effects of
folate on the methylation status of miR-143 further supported the
role of miR-143 in VSMC proliferation. As DNA hypermethylation
inhibits gene expression (35),
the results of the present study demonstrated that aberrant miR-143
DNA hypermethylation was responsible for the downregulation in the
expression of miR-143 in Hcy-induced VSMCs.
It is well supported that altered gene-specific
methylation triggered by abnormal functioning of DNMTs may cause
the transcriptional silencing of cancer-associated genes in human
cancer (36). Therefore, aberrant
gene-specific methylation changes in VSMCs prompted the present
study to investigate whether these changes are associated with
altered expression of DNMTs in VSMCs. DNMT3a is responsible for
setting genomic DNA methylation patterns, a key layer of epigenetic
information (37). It has been
previously demonstrated that miR-143 targets DNMT3a mRNA (30). In order to verify whether DNMT3a
mediates the miR-143 DNA hypermethylation process, the present
study examined the expression of DNMT3a in VSMCs induced by Hcy.
The results showed that the mRNA and protein levels of DNMT3a were
significantly upregulated in the VSMCs, and were downregulated in
the folate group. A similar previous study demonstrated that DNMT3a
mRNA levels was high in breast tumors, accompanied by DNA
hypermethylation (38). In
addition, miR-143 methylation levels in the present study were
decreased following transfection of the VSMCs with the DNMT3a siRNA
plasmid, which further verified that DNMT3a was important in
miR-143 hypermethylation. However, the mechanism responsible for
DNMT3a upregulation remain to be fully elucidated
It has been previously demonstrated that certain
microRNAs targeting DNMTs transcripts lead to the demethylation and
transcriptional activation of numerous protein coding gene
sequences, thereby contributing to gene expression (39). In the present study, the
upregulation of DNMT3a in VSMCs was accompanied by an overall
decrease in the expression of miRNA-143 in the VSMCs induced by
Hcy, which suggested that miR-143 may control the functioning of
DNMT3a. Notably, using bioinformatics analysis, our previous study
found that DNMT3a is one potential target of miR-143 (30). To confirm whether DNMT3a is the
direct target of miR-143 in VSMCs in the present study, pre-miR-143
and a miR-143 inhibitor were co-transfected with the recombinant
plasmid containing the DNMT3a complementary site. The luciferase
activity of DNMT3a were negatively correlated with the levels of
miR-143 levels in the VSMCs transfected with the pre-miR-143 and
inhibitor. Similarly, an inverse correlation was observed between
the expression of miR-143 and DNMT3a in the VSMCs, further
confirming that miR-143 negatively regulated DNMT3a.
In conclusion, the data of the present study
demonstrated for the first time, to the best of our knowledge, that
the upregulation of DNMT3a directly results in the hypermethylation
of miR-143 genes, leading to a downregulation in the expression of
miR-143 expression, accompanied by VSMC proliferation. These
results demonstrate a novel regulatory circuit between DNMT3a and
miR-143 in VSMC proliferation.
Acknowledgments
This study was supported by grants from the National
Natural Science Foundation of China (grant. no. 1260105, 81360053,
81360027 and 81200118), the Ningxia Education Department Scientific
and Technological Project (grant nos. NGY2013054 and NGY2013082)
and the Ningxia Science and Technique Project (grant no.
NZ14120).
References
|
1
|
van den Brandhof WE, Haks K, Schouten EG
and Verhoef P: The relation between plasma cysteine, plasma
homocysteine and coronary atherosclerosis. Atherosclerosis.
157:403–9. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ozkan Y, Ozkan E and Simşek B: Plasma
total homocysteine and cysteine levels as cardiovascular risk
factors in coronary heart disease. Int J Cardio. 82:269–277. 2002.
View Article : Google Scholar
|
|
3
|
Zhang D, Wen X, Wu W, Xu E, Zhang Y and
Cui W: Homocysteine-related hTERT DNA demethylation contributes to
shortened leukocyte telomere length in atherosclerosis.
Atherosclerosis. 231:173–179. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Horvath B, Szapary L, Debreceni L, Feher
G, Kenyeres P, Fulop A, Battyani I and Toth K: Effect of Sclerovit
on endothelial dysfunction, hemorheological parameters, platelet
aggregation, plasma concentration of homocysteine and progression
of atherosclerosis in patients with vascular diseases. Clin
Hemorheol Microcirc. 42:19–28. 2009.PubMed/NCBI
|
|
5
|
Liang Y, Yang X, Ma L, Cai X, Wang L, Yang
C, Li G, Zhang M, Sun W and Jiang Y: Homocysteine-mediated
cholesterol efflux via ABCA1 and ACAT1 DNA methylation in THP-1
monocyte-derived foam cells. Acta Biochim Biophys Sin (Shanghai).
45:220–228. 2013. View Article : Google Scholar
|
|
6
|
Jacob T, Hingorani A and Ascher E:
Evidence for telomerase activation in VSMCs exposed to
hyperglycemic and hyperhomocysteinemic conditions. Angiology.
60:562–568. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kim Y, Han JH, Yun E, Jung SH, Lee JJ,
Song GY and Myung CS: Inhibitory effect of a novel naphthoquinone
derivative on proliferation of vascular smooth muscle cells through
suppression of platelet-derived growth factor receptor β tyrosine
kinase. Eur J Pharmacol. 733:81–89. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Huang J and Kontos CD: Inhibition of
vascular smooth muscle cell proliferation, migration and survival
by the tumor suppressor protein PTEN. Arterioscler Thromb Vasc
Biol. 22:745–751. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yonemitsu Y, Kaneda Y, Tanaka S, Nakashima
Y, Komori K, Sugimachi K and Sueishi K: Transfer of wild-type p53
gene effectively inhibits vascular smooth muscle cell proliferation
in vitro and in vivo. Circ Res. 82:147–156. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang C: MicroRNA and vascular smooth
muscle cell phenotype: New therapy for atherosclerosis? Genome Med.
1:852009. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Rangrez AY, Massy ZA, Metzinger-Le Meuth V
and Metzinger L: MiR-143 and miR-145: Molecular keys to switch the
phenotype of vascular smooth muscle cells. Circ Cardiovasc Genet.
4:197–205. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Dong S, Xiong W, Yuan J, Li J, Liu J and
Xu X: MiRNA-146a regulates the maturation and differentiation of
vascular smooth muscle cells by targeting NF-κB expression. Mol Med
Rep. 8:407–412. 2013.PubMed/NCBI
|
|
14
|
Boucher JM, Peterson SM, Urs S, Zhang C
and Liaw L: The miR-143/145 cluster is a novel transcriptional
target of Jagged-1/Notch signaling in vascular smooth muscle cells.
J Biol Chem. 286:28312–28321. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Saetrom P, Snøve O Jr and Rossi JJ:
Epigenetics and microRNAs. Pediatr Res. 61:17R–23R. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hutnick LK, Golshani P, Namihira M, Xue Z,
Matynia A, Yang XW, Silva AJ, Schweizer FE and Fan G: DNA
hypomethylation restricted to the murine forebrain induces cortical
degeneration and impairs postnatal neuronal maturation. Hum Mol
Genet. 18:2875–2888. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liu K, Wang YF, Cantemir C and Muller MT:
Endogenous assays of DNA methyltransferases: Evidence for
differential activities of DNMT1, DNMT2 and DNMT3 in mammalian
cells in vivo. Mol Cell Biol. 23:2709–2719. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Arand J, Spieler D, Karius T, Branco MR,
Meilinger D, Meissner A, Jenuwein T, Xu G, Leonhardt H, Wolf V and
Walter J: In vivo control of CpG and non-CpG DNA methylation by DNA
methyltransferases. PLoS Genet. 8:e10027502012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Krebs HA, Hems R and Tyler B: The
regulation of folate and methionine metabolism. Biochem J.
158:341–353. 1976. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Toyota M, Suzuki H, Sasaki Y, Maruyama R,
Imai K, Shinomura Y and Tokino T: Epigenetic silencing of
microRNA-34b/c and B-cell translocation gene 4 is associated with
CpG island methylation in colorectal cancer. Cancer Res.
68:4123–4132. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Veeck J and Esteller M: Breast cancer
epigenetics: From DNA methylation to microRNAs. J Mammary Gland
Biol Neoplasia. 15:5–17. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Robaina MC, Mazzoccoli L, Arruda VO, Reis
FR, Apa AG, de Rezende LM and Klumb CE: Deregulation of DNMT1,
DNMT3B and miR-29s in Burkitt lymphoma suggests novel contribution
for disease pathogenesis. Exp Mol Pathol. 98:200–207. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Rao X, Huang X, Zhou Z and Lin X: An
improvement of the 2ˆ(-delta delta CT) method for quantitative
real-time polymerase chain reaction data analysis. Biostat
Bioinforma Biomath. 3:71–85. 2013.PubMed/NCBI
|
|
24
|
Yideng J, Jianzhong Z, Ying H, Juan S,
Jinge Z, Shenglan W, Xiaoqun H and Shuren W: Homocysteine-mediated
expression of SAHH, DNMTs, MBD2 and DNA hypomethylation potential
pathogenic mechanism in VSMCs. DNA Cell Biol. 26:603–611. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ng EK, Tsang WP, Ng SS, Jin HC, Yu J, Li
JJ, Röcken C, Ebert MP, Kwok TT and Sung JJ: MicroRNA-143 targets
DNA methyltransferases 3A in colorectal cancer. Br J Cancer.
101:699–706. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Pawlak K, Mysliwiec M and Pawlak D:
Hyperhomocysteinemia and the presence of cardiovascular disease are
associated with kynurenic acid levels and carotid atherosclerosis
in patients undergoing continuous ambulatory peritoneal dialysis.
Thromb Res. 129:704–9. 2012. View Article : Google Scholar
|
|
27
|
Chistiakov DA, Orekhov AN and Bobryshev
YV: Vascular smooth muscle cell in atherosclerosis. Acta Physiol
(Oxf). 214:33–50. 2015. View Article : Google Scholar
|
|
28
|
Choe N, Kwon JS, Kim JR, Eom GH, Kim Y,
Nam KI, Ahn Y, Kee HJ and Kook H: The microRNA miR-132 targets
Lrrfip1 to block vascular smooth muscle cell proliferation and
neointimal hyperplasia. Atherosclerosis. 229:348–355. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Davis-Dusenbery BN, Chan MC, Reno KE,
Weisman AS, Layne MD, Lagna G and Hata A: downregulation of
Kruppel-like factor-4 (KLF4) by microRNA-143/145 is critical for
modulation of vascular smooth muscle cell phenotype by transforming
growth factor-beta and bone morphogenetic protein 4. J Biol Chem.
286:28097–28110. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhao W, Zhao SP and Zhao YH:
MicroRNA-143/-145 in cardiovascular diseases. Biomed Res Int.
2015:5317402015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Isa Y, Mishima T, Tsuge H and Hayakawa T:
Increase in S-adenosylhomocysteine content and its effect on the
S-adenosylhomocysteine hydrolase activity under transient high
plasma homocysteine levels in rats. J Nutr Sci Vitaminol (Tokyo).
52:479–482. 2006. View Article : Google Scholar
|
|
32
|
Smith DE, Smulders YM, Blom HJ, Popp J,
Jessen F, Semmler A, Farkas M and Linnebank M: Determinants of the
essential one-carbon metabolism metabolites, homocysteine,
S-adenosylmethionine, S-adenosylhomocysteine and folate, in
cerebrospinal fluid. Clin Chem Lab Med. 50:1641–1647. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Sandhu R, Rivenbark AG, Mackler RM, Livasy
CA and Coleman WB: Dysregulation of microRNA expression drives
aberrant DNA hypermethylation in basal-like breast cancer. Int J
Oncol. 44:563–572. 2014.
|
|
34
|
Ma S, Zhang H, Sun W, Gong H, Wang Y, Ma
C, Wang J, Cao C, Yang X, Tian J and Jiang Y: Hyperhomocysteinemia
induces cardiac injury by up-regulation of p53-dependent Noxa and
Bax expression through the p53 DNA methylation in ApoE(−/−) mice.
Acta Biochim Biophys Sin (Shanghai). 45:391–400. 2013. View Article : Google Scholar
|
|
35
|
Magdinier F, Billard LM, Wittmann G,
Frappart L, Benchaïb M, Lenoir GM, Guérin JF and Dante R: Regional
methylation of the 5′ end CpG island of BRCA1 is associated with
reduced gene expression in human somatic cells. FASEB J.
14:1585–1594. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
He M, Fan J, Jiang R, Tang WX and Wang ZW:
Expression of DNMTs and genomic DNA methylation in gastric signet
ring cell carcinoma. Mol Med Rep. 8:942–948. 2013.PubMed/NCBI
|
|
37
|
Holz-Schietinger C, Matje DM and Reich NO:
Mutations in DNA methyltransferase (DNMT3A) observed in acute
myeloid leukemia patients disrupt processive methylation. J Biol
Chem. 287:30941–30951. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Yu Z, Xiao Q, Zhao L, Ren J, Bai X, Sun M,
Wu H, Liu X, Song Z, Yan Y, et al: DNA methyltransferase 1/3a
overexpression in sporadic breast cancer is associated with reduced
expression of estrogen receptor-alpha/breast cancer susceptibility
gene 1 and poor prognosis. Mol Carcinog. 2014.
|
|
39
|
Sun X, He Y, Huang C, Ma TT and Li J: The
epigenetic feedback loop between DNA methylation and microRNAs in
fibrotic disease with an emphasis on DNA methyltransferases. Cell
Signal. 25:1870–1876. 2013. View Article : Google Scholar : PubMed/NCBI
|