Introduction
Lung cancer is the most commonly diagnosed cancer
and leading cause of cancer-associated mortality worldwide,
accounting for the mortality of >1 million individuals each year
(1–3). Non-small cell lung cancer (NSCLC), including
adenocarcinoma and squamous cell carcinoma, is the predominant form
of lung cancer, accounting for 75–80% of all cases (4). Previous evidence revealed that miRNAs
may be involved in lung cancer, and act as oncogenes or tumor
suppressor genes. For example, miR-340 (5), miR-128 (6) and miR-197 (7) were revealed to be deregulated in
NSCLC apoptosis. These studies provided novel insights into lung
cancer biology, and merited further investigation.
Micro (mi)RNAs are small, non-coding RNAs, 18–23
nucleotides in length, which post-transcriptionally regulate gene
expression by base-pairing with target mRNAs in the 3′-untranslated
region (UTR) (8). Emerging
evidence clearly suggests that the deregulation or dysfunction of
miRNAs is associated with crucial biological processes, including
development, differentiation, proliferation and apoptosis (9,10).
miRNAs function as oncogenes or tumor suppressor genes, depending
on the roles of their target genes. Among the miRNAs which are
associated with carcinogenesis, miR-1915-3p is one of the most
important. The dysregulation of miR-1915-3p was reported in various
types of cancer, including prostate cancer (11), renal cell carcinoma (12) and breast cancer (13). miR-1915 regulated the expression of
CD133, Paired box gene 2 and Toll-like receptor 2 in adult renal
progenitor cells (14). miR-1915
suppressed the expression of B-cell lymphoma (Bcl)-2 at the
post-transcriptional level to modulate multidrug resistance by
increasing drug sensitivity in human colorectal carcinoma cells
(15,16). However, miR-1915-3p remains to be
implicated in lung cancer.
In the present study, the role of miR-1915-3p in the
etoposide (VP16)-induced apoptosis of lung cancer cells was
investigated. miR-1915-3p prevented VP16-induced cell apoptosis,
and further investigation revealed that developmentally regulated
GTP-binding protein 2 (DRG2) and pre-B cell leukemia homeobox 2
(PBX2) were direct and functional targets of miR-1915-3p.
Furthermore, DRG2 and PBX2 may partly circumvent the effect of
miR-1915-3p on the apoptosis of H441 and H1650 cells. Therefore,
miR-1915-3p is a putative therapeutic agent for the treatment of
lung cancer.
Materials and methods
Cell culture
NCI-H441 (H441) and NCI-H1650 (H1650) cells
(American Type Culture Collection, Arlington, VA, USA) were
maintained in RPMI-1640 medium (Gibco; Thermo Fisher Scientific,
Inc., Waltham, MA, USA), supplemented with 10% fetal bovine serum
(Beijing Yuanheng Shengma Biotechnology Research Institute,
Beijing, China), 100 U/ml streptomycin (Biological Industries,
Kibbutz Beit-Haemek, Israel) and 100 U/ml penicillin (Biological
Industries), and were maintained at 37°C in a humidified 5%
CO2 incubator. The medium was changed on alternate days
and the cells were split prior to reaching 100% confluence.
Oligonucleotides, plasmids and
transfection
An miR-1915-3p mimic (a chemically synthesized,
double-stranded miRNA) and miR-1915-3p inhibitor were purchased
from GenePharma (Shanghai, China).
The full-length 3′-UTR of DRG2 or PBX2 was subcloned
into the pIS0 luciferase plasmid (17) to generate pIS0-DRG2-3′-UTR or
pIS0-PBX2-3′-UTR, respectively. Mutant constructs of DRG2 and
PBX2-3′-UTR, termed pIS0-DRG2-3′-UTR-mut and pIS0-PBX2-3′-UTR-mut,
respecitvely, which bore a replacement of three nucleotides within
the core binding sites of DRG2 or PBX2-3-UTR, were synthesized
using mutant PCR primers. The primer pairs were as follows:
pIS0-DRG2-3′-UTR-forward (F), 5′-TTGAGCTCCCAGCACCAAGTACAGTC-3′ and
-reverse (R), 5′-GCTCTAGAAAGGAGAGCCAGGAGAAC-3′;
pIS0-DRG2-3′-UTR-mut-F, 5′-CCTCGTCTCCAGTGGGAGGTGG-3′ and -R,
5′-CCACCTCCCACTGGAGACGAGG-3′; pIS0-PBX2-3′-UTR-F,
5′-TTGAGCTCCGGACGGCTTACTTACCT-3′ and -R,
5′-GCTCTAGACTTCACTGCCTCCACATC-3′; pIS0-PBX2-3′-UTR-mut-F,
5′-GAAAAAGAAACCAGTGGATCCATCT-3′ and -R,
5′-AGATGGATCCACTGGTTTCTTTTTC-3′.
The cells were seeded into 6-well plates at
2×105 cells/well. When the cell density reached 60–70%,
the cells were trans-fected with DNA plasmid and oligonucleotide
using Lipo-fectamine 2000 (Invitrogen; Thermo Fisher Scientific,
Inc.), according to the manufacturer's instructions.
Prediction of miRNA targets
In order to investigate the predicted target genes,
the TargetScan program (http://www.targetscan.org/) and the miRbase program
(www.mirbase.org) were used.
RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
The total RNA was extracted using Invitrogen TRIzol
reagent (Thermo Fisher Scientific, Inc.). The RT was performed
using a FastQuant RT kit [TianGen Biotech (Beijing) Co., Ltd,
Beijing, China]. miR-1915-3p was reverse-transcribed using a
stem-loop primer
(5′-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGCCCGCCGC-3′). The RT of
DRG2/PBX2 mRNA was performed using a FastQuant RT kit [TianGen
Biotech (Beijing) Co., Ltd], according to the manufacturer's
protocol. RT-qPCR was performed using a SuperReal PreMix Plus kit
[TianGen Biotech (Beijing) Co., Ltd]. U6 small nuclear RNA and
β-actin mRNA were used as internal controls for miR-1915-3p and
DRG2/PBX2 mRNA, respectively. All reactions were performed in
triplicate. The primers used were as follows: DRG2-F,
5′-GCTGAACCTGGACTATCTG-3′ and -R, 5′-GAATGATGGCGTCTGTGA-3′; PBX2-F,
5′-CCC ATG TCA TGA ACC TGC TG-3′ and -R, 5′-GCG CTG AAC TTT CGA TGG
AT-3′. The relative fold changes were calculated using the
−2∆∆Cq method, and β-actin was used as an internal
control.
Luciferase assay
The cells were cultured in 96-well plates and
transiently co-transfected with the miR-1915-3p mimic and the pIS0
vectors using Lipofectamine 2000 reagent (Invitrogen; Thermo Fisher
Scientific, Inc.). A scrambled sequence
(5′-AACCACUCAACUUUUUCCCAAdTdT-3′) was used as a negative control.
Following an incubation for 48 h, luciferase activity was measured
using a dual luciferase reporter assay system (Promega, Madison,
WI, USA). pRL-TK Renilla was used as an internal control.
Three independent experiments were performed, and all reactions
were performed in triplicate.
Flow cytometric analysis (FCM)
The cells were seeded into 6-well plates at a
density of 5×104 cells/well, and were transfected with
the miR-1915-3p mimic or miR-1915-3p inhibitor for 24 h, starved
overnight, and treated with etoposide for 48 h. The apoptotic cells
were detected using the Annexin V-FITC Apoptosis Detection kit (BD
Biosciences, San Jose, CA, USA) and analyzed on a FACSCalibur™ flow
cytometer (BD Biosciences) using CellQuest software version
3.3.
Western blotting
The cellular proteins were extracted following
treatments, as previously described (18). The protein (20 µg) was
loaded into each lane of 8% polyacryl-amide gels. The proteins were
separated by electrophoresis and the proteins were blotted onto
polyvinylidene difluoride membranes (Amersham Pharmacia Biotech,
St. Albans, UK) by electrophoretic transfer. The membrane was
incubated with primary antibodies goat anti-DRG2 (1:750; cat. no.
sc-164233; Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA),
rabbit anti-PBX2 (1:1,000; cat. no. sc-980; Santa Cruz
Biotechnology, Inc.) and rabbit anti-β-actin (1:3,000; cat. no.
4970; Cell Signaling Technology, Inc., Beverly, MA, USA), which was
used as a loading control.
Statistical analysis
The data are expressed as the mean ± standard
deviation from at least three separate experiments, and Student's
t-test was performed using SPSS 17.0 software (SPSS, Inc., Chicago,
IL, USA). P<0.05 was considered to indicate a statistically
significant difference.
Results
miR-1915-3p is involved in lung cancer
cell apoptosis
Etoposide (VP16) is known to exert its cancer cell
killing effects through induction of apoptosis (19). VP16 induces apoptosis in a number
of cancer cell types (20–23), and has been used for the treatment
of a wide variety of cancer types (24). In the present study, a marked
increase in the levels of apoptosis were detected in H441 and H1650
cells following treatment with VP16 (Fig. 1A and B). RT-qPCR was subsequently
performed to assess whether any changes in the level of miR-1915-3p
occurred. The expression of miR-1915-3p was markedly down-regulated
during VP16-induced lung cancer cell apoptosis (Fig. 1C). The reduced expression level of
miR-1915-3p during lung cancer apoptosis suggested that this miRNA
may exert an antiapoptotic effect. To examine this hypothesis, the
effects of transient transfection of the miR-1915-3p mimic, or
addition of the miR-1915-3p inhibitor to lung cancer H441 cells,
were investigated. Following transfection, the cells were deprived
of serum overnight, and were subsequently treated with VP16 for 48
h. FCM was performed to detect the number of apoptotic cells
present. Overexpression of the miR-1915-3p mimic markedly inhibited
VP16-induced apoptosis (Fig. 1D).
By contrast, overexpression of the miR-1915-3p inhibitor elicited
the opposite effect, promoting VP16-induced apoptosis (Fig. 1D). A similar phenomenon was also
observed in the lung cancer H1650 cells (Fig. 1E). Therefore, these results
indicated that miR-1915-3p was associated with lung cancer cell
apoptosis.
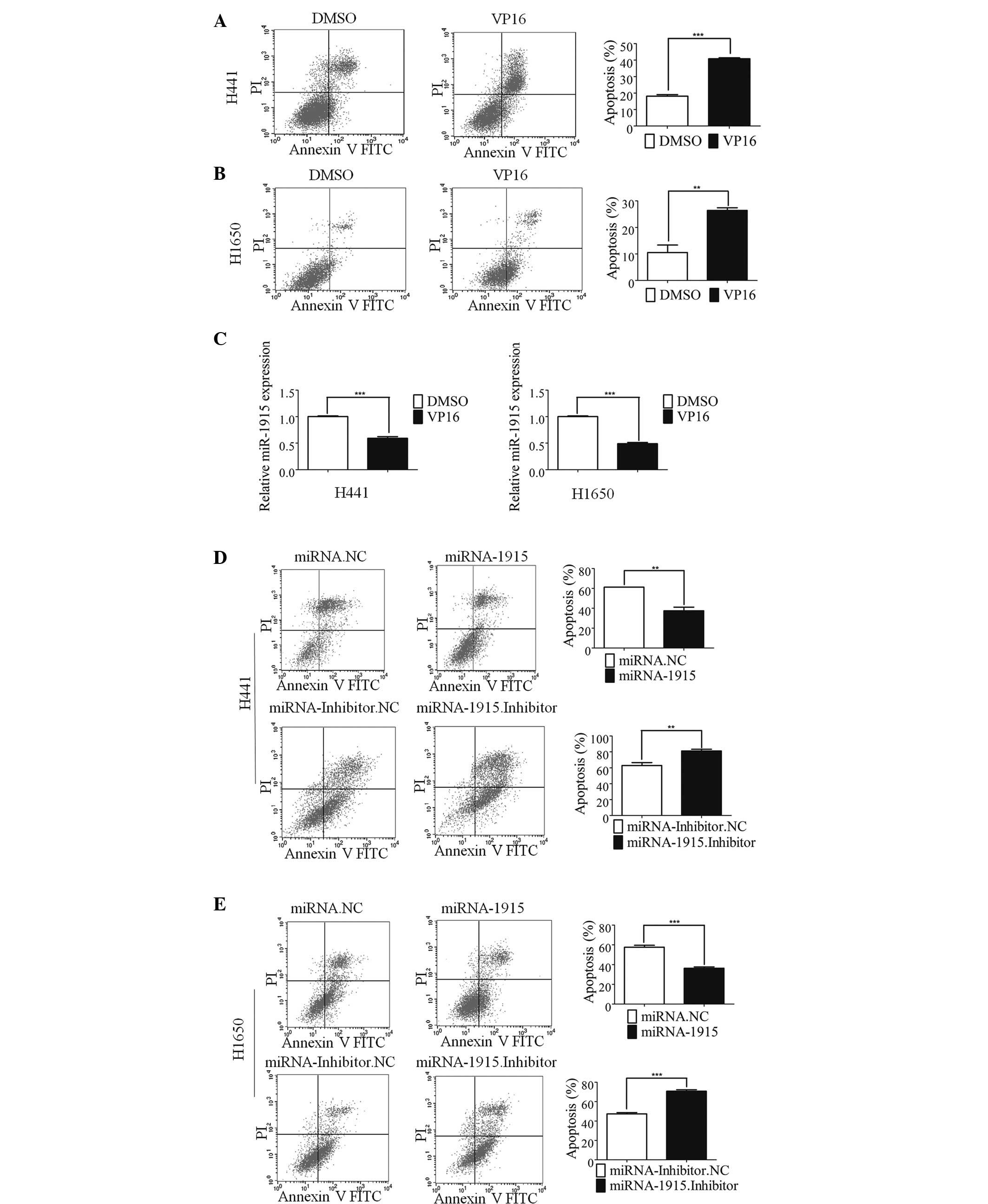 | Figure 1miR-1915 is involved in lung cancer
cell apoptosis. (A) The apoptosis of the H441 cells was induced by
treatment with 100 µM etoposide (VP16) for 48 h. The
histogram shows the percentage of apoptotic cells determined by
FCM, and the error bars denote the mean ± standard deviation. (B)
The results of FCM are shown, revealing the apoptosis of the H1650
cells following treatment with 100 µM VP16. The histogram
shows the percentage of apoptotic cells determined by FCM, and the
error bars denote the mean ± standard deviation. (C) The histogram
shows the expression of miRNA-1915-3p in the H441 (left) and H1650
cells (right) at 48 h following treatment with 100 µM VP16
treatment. (D) The apoptosis of the H441 cells was induced by
treatment with VP16 for 48 h following 24 h transfection with
miR-1915-3p mimic. The histogram shows the percentage of apoptotic
cells determined by FCM, and the error bars denote the mean ±
standard deviation. (E) The apoptosis of H1650 cells induced by
treatment with VP16 following miR-1915-3p mimic transfection for 24
h is shown. The histogram shows the percentage of apoptotic cells
determined by FCM, and the error bars denote the mean ± standard
deviation (**P<0.01 and ***P<0.001,
compared with the control, using Student's t-test). DMSO, dimethyl
sulfoxide; FCM, flow cytometric analysis; FITC, fluorescein
isothiocyanate; miRNA, microRNA; NC, negative control; PI,
propidium iodide. |
DRG2 and PBX2 are direct targets of
miR-1915-3p
To assess the molecular mechanism of miR-1915-3p in
the regulation of apoptosis, putative miR-1915-3p targets were
predicted using the target prediction programs, TargetScan, miRanda
and miRbase. DRG2 and PBX2 were identified as potential targets of
miR-1915-3p. The 3′-UTR of the DRG2 and PBX2 mRNA contained a
complementary site for the seed region of miR-1915-3p (Fig. 2A). To determine whether DRG2 and
PBX2 were regulated by miR-1915-3p through direct binding to their
3′-UTR sequences, a human DRG2/PBX2 3′-UTR fragment containing
wild-type (wt) or mutant miR-1915-3p binding sequence were cloned
downstream of the firefly lucif-erase reporter gene.
Co-transfection of the luciferase reporter,
pIS0-DRG2/PBX2-3′-UTR-wt and miR-1915-3p, into the lung cancer
H1650 cells produced a 50–80% decrease in the luciferase activity
compared with the negative control (Fig. 2B). This suppressive effect was
rescued by the three nucleotide substitutions
(pIS0-DRG2/PBX2-3ʼUTR-mutant) in the core binding sites, as shown
in Fig. 2B (upper panels). A
similar effect was identified for the lung cancer H441 cells
(Fig. 2B, lower panels).
 | Figure 2DRG2 and PBX2 are direct targets of
miR-1915-3p. (A) The miR-1915-3p targeting site resides at nt
38–44 of the DRG2-3′UTR and nt 701–708 of the
PBX2-3′-UTR. The upper panels are the sequence alignment of
miR-1915-3p with binding sites on the DRG2- and PBX2-3′-UTR. The
lower panel is a diagram of the luciferase reporter plasmids,
including the plasmids with the full-length DRG2-3′-UTR and
PBX2-3′-UTR insert (pIS0-DRG2-3′UTR and pIS0-PBX2-3′UTR,
respectively) and plasmids with a mutant DRG2-3′-UTR and
PBX2-3′-UTR (pIS0-DRG2-3′UTR-MUT and pIS0-PBX2-3′UTR-MUT,
respectively), which harbored a replacement of three nts within the
miR-1915-3p-binding site. (B) H1650 (upper panels) and H441 (lower
panels) cells were transfected with the miR-1915-3p mimic (20 nM)
or the plasmids pIS0-DRG2/PBX2-3′-UTR or pIS0-DRG2/PBX2-3′-UTR-mut.
pRL-SV40 Renilla was used for normalization of the
transfection efficiency. Following an incubation for 48 h, the
luciferase activities were measured. (C) The expression levels of
the DRG2 and PBX2 mRNAs in H1650 (left panel) or H441 (right panel)
cell lines, treated as described above, were measured using
RT-qPCR. β-Actin was used as internal control. (D) Western blotting
was used to detect the expression of DRG2 and PBX2 proteins
following transfection of the miRNA-1915-3p mimic (20 nM) or the
miRNA-1915-3p inhibitor (40 nM) into the H1650 (left panels) or
H441 (right panels) cells. *P<0.05;
**P<0.01; ***P<0.001, compared with the
NC experiments. DRG2, developmentally regulated GTP-binding protein
2; NC. negative control; nt, nucleotide; PBX2, pre-B cell leukemia
homeobox 2; UTR, untranslated region; miRNA, microRNA. |
The effect of miRNA-1915 on the endogenous
expression of DRG2/PBX2 was further examined. Marked decreases in
the endogenous mRNA (Fig. 2C) and
protein (Fig. 2D) expression
levels of DRG2 and PBX2 were observed in the H441 and H1650 cells
transfected with the miR-1915-3p mimic, whereas transfection with
the miR-1915-3p inhibitor induced a marked increase in the protein
expression levels of DRG2/PBX2 (Fig.
2D). These results suggested that miR-1915-3p inhibited the
expression of DRG2/PBX2 at the post-transcriptional level by
directly targeting the 3′-UTRs of the DRG2/PBX2 mRNA.
Overexpression of DRG2/PBX2 induces
apoptosis of the H441 and H1650 lung cancer cells
Since miR-1915-3p decreased the expression of DRG2
and PBX2 and inhibited apoptosis, consequently, the association of
DRG2 or PBX with the process of apoptosis in the lung cancer cells
was investigated. The H441 cells were transfected with a plasmid
harboring DRG2 or PBX2 and deprived of serum overnight, followed by
treatment with VP16 for 48 h. FCM was performed to assess the
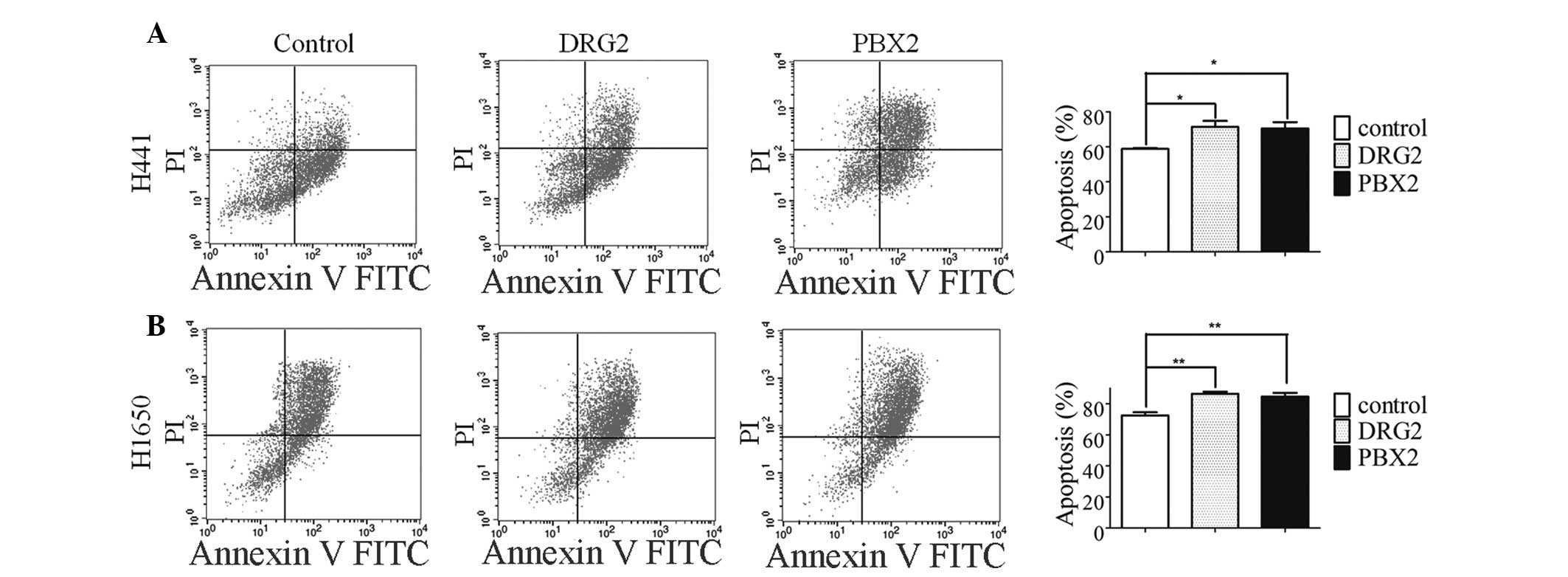
numbers of apoptotic cells. The overexpression of DRG2 or PBX2
significantly promoted VP16-induced apoptosis (Fig. 3A). A similar phenomenon was
observed in the H1650 lung cancer cells (Fig. 3B). The identification of DRG2 and
PBX2 as novel miR-1915-3p target genes may explain, at least in
part, the molecular mechanism of miR-1915-3p.
miRNA-1915 regulates apoptosis through
DRG2 and PBX2
Subsequently, whether DRG2 and PBX2 were involved in
apoptosis regulated by miRNA-1915-3p was investigated. H1650 cells
were co-transfected with the miR-1915-3p mimic and pcDNA3-DRG2 or
pcDNA3-PBX2, which encoded the full-length coding region of
DRG2/PBX2, although they lacked the 3′-UTR of the DRG2/PBX2 mRNA.
The results indicated that overexpression of miR-1915-3p alone
decreased the number of VP16-induced apoptotic cells, whereas
co-expression with DRG2 or PBX2 effectively reversed the change
(Fig. 4A). A similar phenomenon
was observed with the H441 cells (Fig.
4B). These results suggested that the apoptotic involvement of
miR-1915-3p, at least in part, was dependent on its role in
regulating the expression levels of DRG2 and PBX2.
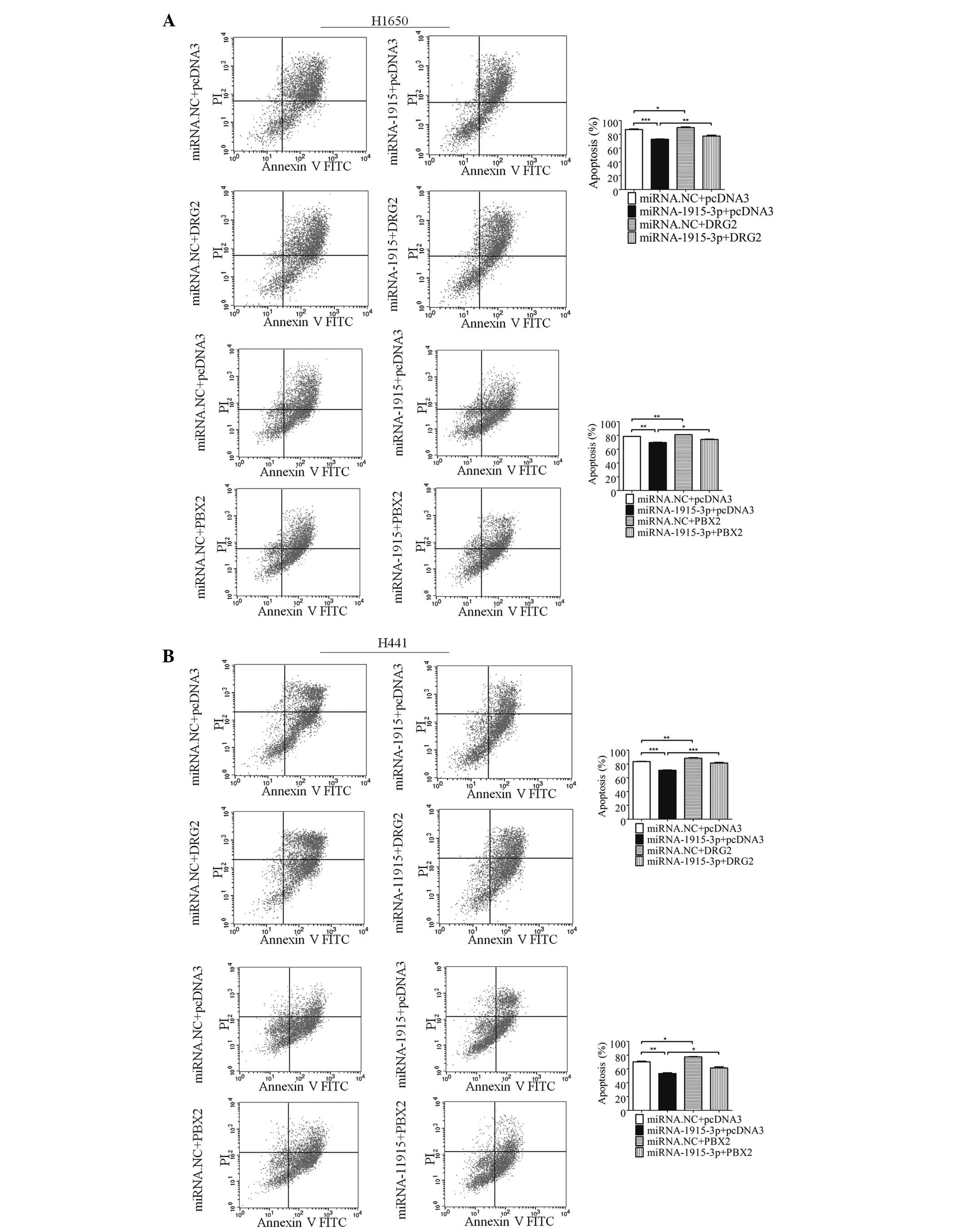 | Figure 4miR-1915-3p regulates apoptosis
through DRG2 and PBX2. (A) Apoptosis of the H1650 cells transfected
with the miRNA-1915-3p mimic, and/or combined with DRG2 or PBX2 for
48 h was determined using FCM (left). The error bars in the
histograms on the right denote the mean ± standard deviation. (B)
Apoptosis of the H441 cells at 48 h following transfection of the
miRNA-1915-3p mimic, and/or combined with DRG2 or PBX2, was
determined by FCM (left). The error bars in the histograms on the
right denote the mean ± standard deviation (*P<0.05,
**P<0.01 and ***P<0.001, compared with
the miRNA. NC+pcDNA3 control, or the miRNA-1915-3p+pcDNA3
experiments). PI, propidium iodide; FITC, fluorescein
isothiocyanate; FCM, flow cytometric analysis; DRG2,
developmentally regulated GTP-binding protein 2; FCM, flow
cytometric analysis; NC, negative control; PBX2, pre-B cell
leukemia homeobox 2; miRNA, microRNA. |
Discussion
The association of a series of miRNAs with cellular
apoptosis has been previously experimentally verified (25–27).
In the present study, it was demonstrated that miR-1915-3p was a
candidate regulator of lung cancer cell apoptosis. This finding
therefore expanded the list of miRNAs which are known to be
involved in regulating apoptosis.
The experiments performed in the present study
demonstrated that DRG2 and PBX2 were targets of miR-1915-3p. By
interacting directly with the 3′-UTRs of the DRG2 and PBX2 mRNA,
miR-1915-3p regulated the expression of DRG2 and PBX2 at the
post-transcriptional level. The overex-pression of miR-1915-3p
eliminated VP16-induced apoptosis and led to a marked decrease in
the endogenous protein expression of DRG2 and PBX2. DRG2 and PBX2
were therefore associated with apoptosis.
In previously published studies, the overexpression
of DRG2 inhibited doxorubicin-induced apoptosis in hepatocellular
carcinoma cells (28) and reduced
the sensitivity of Jurkat cells to the antimitotic agent,
nocodazole (29), although it is
unclear whether DRG2 exerts a role in Jurkat cell apoptosis
(30). PBX2 is a transcriptional
activator, which binds to the T-cell leukemia homeobox protein 1
promoter, and a high level of expression serves as an independent,
negative prognostic indicator for gastric cancer and esophageal
squamous cell carcinoma (31).
However, the roles of DRG2 and PBX2 in lung cancer apoptosis remain
to be elucidated. In the present study, it has been demonstrated
that the overexpression of DRG2 and PBX2 may markedly promote the
VP16-induced apoptosis of lung cancer cells.
In conclusion, the present study demonstrated for
the first time, to the best of our knowledge, that the levels of
miR-1915-3p are significantly downregulated during the apop-tosis
of lung cancer cells. The overexpression of miR-1915-3p markedly
inhibited VP16-induced cell apoptosis. DRG2 and PBX2 were
identified as direct and functional targets of miR-1915-3p.
Finally, it was demonstrated that miR-1915 regulates apoptosis via
DRG2 and PBX2. These findings may lead to novel therapeutic options
for treating human lung cancer.
Acknowledgments
The present study was supported by the National
Natural Science Foundation of China (nos. 81070424 and
81272303).
References
|
1
|
Siegel R, Ma J, Zou Z and Jemal A: Cancer
statistics, 2014. CA Cancer J Clin. 64:9–29. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ferlay J, Shin HR, Bray F, Forman D,
Mathers C and Parkin DM: Estimates of worldwide burden of cancer in
2008: GLOBOCAN 2008. Int J Cancer. 127:2893–2917. 2010. View Article : Google Scholar
|
|
3
|
Spiro SG and Silvestri GA: One hundred
years of lung cancer. Am J Resp Crit Care Med. 17:2523–2529.
2005.
|
|
4
|
Harmsma M, Schutte B and Ramaekers FC:
Serum markers in small cell lung cancer: Opportunities for
improvement. Biochim Biophys Acta. 1836:255–272. 2013.PubMed/NCBI
|
|
5
|
Fernandez S, Risolino M, Mandia N, Talotta
F, Soini Y, Incoronato M, Condorelli G, Banfi S and Verde P:
miR-340 inhibits tumor cell proliferation and induces apoptosis by
targeting multiple negative regulators of p27 in non-small cell
lung cancer. Oncogene. 34:3240–3250. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hu J, Cheng Y, Li Y, Jin Z, Pan Y, Liu G,
Fu S, Zhang Y, Feng K and Feng Y: microRNA-128 plays a critical
role in human non-small cell lung cancer tumourigenesis,
angiogenesis and lymphangiogenesis by directly targeting vascular
endothelial growth factor-C. Eur J Cancer. 50:2336–2350. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Fiori ME, Barbini C, Haas TL, Marroncelli
N, Patrizii M, Biffoni M and De Maria R: Antitumor effect of
miR-197 targeting in p53 wild-type lung cancer. Cell Death Differ.
21:774–782. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Harfe BD: MicroRNAs in vertebrate
development. Curr Opin Genet Dev. 15:410–415. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
He HC, Han ZD, Dai QS, Ling XH, Fu X, Lin
ZY, Deng YH, Qin GQ, Cai C, Chen JH, et al: Global analysis of the
differentially expressed miRNAs of prostate cancer in Chinese
patients. BMC Genomics. 14:7572013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
White NM, Khella HW, Grigull J, Adzovic S,
Youssef YM, Honey RJ, Stewart R, Pace KT, Bjarnason GA, Jewett MA,
et al: miRNA profiling in metastatic renal cell carcinoma reveals a
tumour-suppressor effect for miR-215. Br J Cancer. 105:1741–1749.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chen L, Li Y, Fu Y, Peng J, Mo MH,
Stamatakos M, Teal CB, Brem RF, Stojadinovic A, Grinkemeyer M, et
al: Role of deregulated microRNAs in breast cancer progression
using FFPE tissue. PLoS One. 8:e542132013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sallustio F, Serino G, Costantino V, Curci
C, Cox SN, De Palma G and Schena FP: miR-1915 and miR-1225-5p
regulate the expression of CD133, PAX2 and TLR2 in adult renal
progenitor cells. PLoS One. 8:e682962013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Nakazawa K, Dashzeveg N and Yoshida K:
Tumor suppressor p53 induces miR-1915 processing to inhibit Bcl-2
in the apoptotic response to DNA damage. FEBS J. 281:2937–2944.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Xu K, Liang X, Cui D, Wu Y, Shi W and Liu
J: miR-1915 inhibits Bcl-2 to modulate multidrug resistance by
increasing drug-sensitivity in human colorectal carcinoma cells.
Mol Carcinog. 52:70–78. 2013. View
Article : Google Scholar
|
|
17
|
Yekta S, Shih IH and Bartel DP:
MicroRNA-directed cleavage of HOXB8 mRNA. Science. 304:594–596.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Dong Y, Xiong M, Duan L, Liu Z, Niu T, Luo
Y, Wu X, Xu C and Lu C: H2AX phosphorylation regulated by p38 is
involved in Bim expression and apoptosis in chronic myelogenous
leukemia cells induced by imatinib. Apoptosis. 19:1281–1292. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Pommier Y, Leo E, Zhang H and Marchand C:
DNA topoisomerases and their poisoning by anticancer and
antibacterial drugs. Chem Biol. 17:421–433. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Cui X, Choi HK, Choi YS, Park SY, Sung GJ,
Lee YH, Lee J, Jun WJ, Kim K, Choi KC, et al: DNAJB1 destabilizes
PDCD5 to suppress p53-mediated apoptosis. Cancer Lett. 357:307–315.
2015. View Article : Google Scholar
|
|
21
|
Park JH, Lee SW, Yang SW, Yoo HM, Park JM,
Seong MW, Ka SH, Oh KH, Jeon YJ and Chung CH: Modification of DBC1
by SUMO2/3 is crucial for p53-mediated apoptosis in response to DNA
damage. Nat Commun. 5:54832014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Okamoto S, Jiang Y, Kawamura K, Shingyoji
M, Tada Y, Sekine I, Takiguchi Y, Tatsumi K, Kobayashi H, Shimada
H, et al: Zoledronic acid induces apoptosis and S-phase arrest in
mesothelioma through inhibiting Rab family proteins and
topoisomerase II actions. Cell Death Dis. 5:e15172014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Liu J, Uematsu H, Tsuchida N and Ikeda MA:
Essential role of caspase-8 in p53/p73-dependent apoptosis induced
by etoposide in head and neck carcinoma cells. Mol Cancer.
10:952011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hande KR: Etoposide: Four decades of
development of a topoisomerase II inhibitor. Eur J Cancer.
34:1514–1521. 1998. View Article : Google Scholar
|
|
25
|
Zhang Y, Geng L, Talmon G and Wang J:
MicroRNA-520g confers drug resistance by regulating p21 expression
in colorectal cancer. J Biol Chem. 290:6215–6225. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Adi Harel S, Bossel Ben-Moshe N, Aylon Y,
Bublik DR, Moskovits N, Toperoff G, Azaiza D, Biagoni F, Fuchs G,
Wilder S, et al: Reactivation of epigenetically silenced miR-512
and miR-373 sensitizes lung cancer cells to cisplatin and restricts
tumor growth. Cell Death Differ. 22:1328–1340. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yang CH, Pfeffer SR, Sims M, Yue J, Wang
Y, Linga VG, Paulus E, Davidoff AM and Pfeffer LM: The oncogenic
microRNA-21 inhibits the tumor suppressive activity of FBXO11 to
promote tumorigenesis. J Biol Chem. 290:6037–6046. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chen J, Shen BY, Deng XX, Zhan Q and Peng
CH: SKP1-CULLIN1-F-box (SCF)-mediated DRG2 degradation facilitated
chemotherapeutic drugs induced apoptosis in hepatocellular
carcinoma cells. Biochem Biophys Res Commun. 420:651–655. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Song H, Kim SI, Ko MS, Kim HJ, Heo JC, Lee
HJ, Lee HS, Han IS, Kwack K and Park JW: Overexpression of DRG2
increases G2/M phase cells and decreases sensitivity to
nocodazole-induced apoptosis. J Biochem. 135:331–335. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ko MS, Lee UH, Kim SI, Kim HJ, Park JJ,
Cha SJ, Kim SB, Song H, Chung DK, Han IS, et al: Overexpression of
DRG2 suppresses the growth of Jurkat T cells but does not induce
apoptosis. Arch Biochem Biophys. 422:137–144. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Qiu Y, Song B, Zhao G, Deng B, Makino T,
Tomita Y, Wang J, Luo W, Doki Y, Aozasa K, et al: Expression level
of Pre B cell leukemia homeobox 2 correlates with poor prognosis of
gastric adenocarcinoma and esophageal squamous cell carcinoma. Int
J Oncol. 36:651–663. 2010.PubMed/NCBI
|