Introduction
Osteosarcoma is the most common type of malignant
bone tumor in adolescents and young adults. In ~75% of cases,
patients suffering from osteosarcoma are aged between 15–25 years
old, with a median onset age of 16 years old and a male
predominance (1). Pain and
swelling of the soft tissues are the most common symptoms in
patients with osteosarcoma (2).
Histologically, osteosarcoma is ascribed to the proliferation of
malignant spindle cells and is characterized by osteoid, which is
directly produced by sarcoma cells (3). However, although current
understanding of the histological and clinical manifestations of
osteosarcoma is increasing, knowledge regarding the onset of
osteosarcoma remains limited.
Previous reports identifying fibroblast growth
factor receptors (FGFRs) have significantly improved current
understanding of human tumorigenesis (4–6).
FGFRs are transmembrane tyrosine kinase receptors, which belong to
the immunoglobulin (Ig) superfamily (7). FGFRs are known to be comprised of
four members in humans; FGFR1, FGFR2, FGFR3 and FGFR4 (7). Structurally, the prototypical FGFR
monomer consists of three domains: An extracellular domain, which
mediates FGF binding; a transmembrane domain; and an intracellular
tyrosine kinase domain (7). The
binding of FGFs ligands to FGFRs directly induces receptor
dimerization and finally activates FGFRs kinase activities, leading
to initiation of the intracellular signaling network (7). Increasing evidence indicates that
alteration of the FGF-FGFR signaling cascade may lead to cancer and
is involved in organ development, tumor cell proliferation and
metastasis (6,8–11).
At present, three alterations have been identified as the
predominant mechanisms that contribute to FGFR-mediated human
tumorigenesis, including chromosomal translocations (12–14),
receptor gene amplification (15–17)
and FGFR-activating mutations (18,19).
FGFR1, the first member of the FGFR family, has been
investigated predominantly in the process of human tumori-genesis.
Of note, FGFR1 overexpression is common in multiple types of tumor.
A previous study demonstrated that, in breast cancer, FGFR1
amplification was one of the most common changes and accounted for
10% of breast cancer cases (20).
Evidence has also revealed that the upregulation of FGFR1 increases
cell proliferative ability, whereas its downregulation stimulates
apoptosis in breast cancer (21).
In addition, a previous study reported the existence of focal
amplification of FGFR1 in non-small cell lung cancer and in 21% of
lung adenocarcinoma cases (22).
Furthermore, the number of FGFR1 copies has been identified as an
independent prognostic factor in non-small cell lung cancer
(23), and the FGFR inhibitor,
ponatinib, can suppress the growth of non-small cell lung cancer
cells exhibiting a high expression level of FGFR1 (24). FGFR1 has also found to be
upregulated in prostate cancer (25), pancreatic ductal adenocarcinoma
(26), oral squamous cell
carcinoma (27), bladder cancer
(28), ovarian cancer (29) and sarcoma (30). Although high expression levels of
FGFR1 have been observed in a broad spectrum of types of cancer,
its role in human bone diseases remains to be elucidated. To the
best of our knowledge, FGFR1 has only been reported to be
associated with fracture non-union (31).
The present study aimed to investigate the
expression profile of FGFR1 in osteosarcoma and determine the
possible mechanisms underlying FGFR-mediated osteosarcoma
development, using high-throughput tissue microarray analysis.
Furthermore, the role of FGFR1 in osteosarcoma MG63 cell
proliferation was examined.
Materials and methods
Reagents
FGFR1 cDNA was amplified from the human genome by
polymerase chain reaction (PCR) and the amplified fragments were
digested with HindIII and XhoI (Takara Biotechnology
Co., Ltd., Dalian, China) and were inserted into the HindIII and
XhoI sites of the pcDNA3.1-Flag vector (Invitrogen Life
Technologies, Carlsbad, CA, USA), according to the manufacturer's
instructions. Sequences were verified using DNA electrophoresis and
sequencing. Dulbecco's modified Eagle's medium (DMEM; Corning
Incorporated, New York, NY, USA), dimethylsulfoxide (DMSO;
Sigma-Aldrich, St. Louis, MO, USA) and fetal bovine serum (FBS;
Thermo Fisher Scientific, Waltham, MA, USA) were all purchased from
commercial sources, as described. The anti-FGFR1 and actin
antibodies were purchased from Santa Cruz Biotechnology, Inc.
(Santa Cruz, CA, USA). Primary antibodies against CDK1 and CDK2
were obtained from Abcam (Cambridge, MA, USA).
Cell line and transfection
The MG63 human osteosarcoma cell line was obtained
from Shanghai Institute of Biological Sciences, Chinese Academy of
Sciences (Shanghai, China). The cells were cultured in DMEM with
10% FBS at 37°C in a 5% CO2 humidity-controlled
incubator. The medium was refreshed every 2 days. The Flag-FGFR1
plasmid, which was constructed using the pcDNA3.1-basic vector, was
transfected into the MG63 cells (1×106 cells/6 cm dish)
prior to cell confluence reaching 80%. Lipofectamine 2000
(Invitrogen Life Technologies) was used as the transfection regent.
Transfection efficiency was verified using a fluorescence
microscope (DMI3000 B; Leica Microsystems GmbH, Wetzlar, Germany)
and subsequent western blotting.
Patient samples and immunohistochemisty
(IHC)
A total of 65 patients who underwent surgery at The
Sixth People's Hospital Affiliated to Shanghai Jiaotong University
(Shanghai, China) between 2012 and 2014 were chosen subsequent to
histological verification of osteosarcoma. None of the 65 patients
received preoperative chemotherapy or radiotherapy. The examined
population consisted of 65 patients (41 men and 24 women) with a
median age of diagnosis of 51 years old (range, 33–75 years). The
tissue, which was separate from the edge of tumor foci and was
histologically judged to be free from adenocarcinoma cells, was
used as a normal tissue control. All tissue samples were fixed in
4% formalin immediately subsequent to removal and were embedded in
paraffin for immunohistochemical staining. The study was approved
by the Ethics Committee of Shanghai Jiaotong University, and all
patients provided informed consent.
IHC analysis was performed using osteosarcoma
tissues and paired normal bone tissues. Paraffin-embedded tissues
were first cut into 4 µm-thick sections, followed by
deparaffinization and rehydration with xylene and ethanol. The
sections were then subjected to antigen retrieval in 0.1 M citrate
acid buffer (Sangon Biotech Co., Ltd., Shanghai, China). Following
washing in phosphate-buffered saline (PBS) three times, the
sections were incubated with primary antibodies overnight at 4°C.
Following incubation, the sections were incubated with
horseradish-peroxidase-labeled secondary antibody for 1 h at room
temperature. Following washing in PBS three times, the sections
were visualized using diaminobenzidine and images were captured
(DM13000 B; Leica Microsystems GmbH).
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA were isolated from samples from 65
patients using TRIzol reagent (Sigma-Aldrich). The cDNA (500 ng)
was then prepared using an AMV Reverse Transcriptase synthesis kit
(Promega Corporation, Madison, WI, USA), according to the
manufacturer's instructions. qPCR was performed using a QuantiTect
SYBR Green PCR kit (Qiagen, Shanghai, China). The qPCR cycle
conditions were as follows: 95°C for 30 sec 38 cycles of 95°C for 5
sec and 60°C for 34 sec. The amplification specificity was
evaluated by melting curve analysis. The relative mRNA levels were
determined using the 2−ΔCT (CT; cycle threshold)
formula, where ΔCT = CT (target gene) − CT (actin). All assays were
performed in triplicate. β-actin was selected as the internal
control. The following primers were used: Forward
5′-CTAAGATCCCGTCCATCGCC-3′ and reverse
5′-GGAGCCCAGCACTTTGATCT-3′.
Western blot analysis
Following culture for 24 h, the MG63 cells were
lysed using radioimmunoprecipitation assay buffer (Sigma-Aldrich),
containing 150 mM NaCl, 1% sodium deoxycholate, 0.1% SDS, 1% Triton
X-100, 50 mM Tris-HCl (pH 7.5) and 2 mM EDTA), on ice for 20 min.
Following centrifugation at 4°C for 20 min (12,000 × g), the
supernatant was collected. Protein concentration was determined
using a Bicinchoninic Acid Assay kit (Pierce Biotechnology,
Rockford, IL, USA). Equal quantities of protein extract (40
µg) were loaded into each lane of a 12% SDS-PAGE gel (Sangon
Biotech Co., Ltd.) for separation, and transferred onto
polyvinylidene difluoride membranes (Bio-Rad Laboratories, Inc.,
Hercules, CA, USA). Following antigen blocking in Tris-buffered
saline with 0.1% Tween 20, supplemented with 5% skim milk for 1 h,
the membranes were incubated with the following primary antibodies
overnight at 4°C: Monoclonal rabbit anti-EGFR1 (1:1,000; cat. no.
ab32152; Epitomics, Inc., Burlingame, CA, USA) and monoclonal
rabbit anti-CDK1 (1:1,000; cat. no. 5524; Cell Signaling
Technology, Inc., Danvers, MA, USA). Subsequently, the membranes
were incubated with the goat anti-rabbit IgG (1:5,000; Santa Cruz
Biotechnology, Inc.) secondary antibodies for 1 h at room
temperature and were processed for electrochemiluminescent
detection using an ECL system (Pierce Biotechnology).
Microarray analysis
To investigate the possible pathways contributing to
the onset of FGFR1-mediated osteosarcoma, the present study
performed microarray analysis in osteosarcoma samples. The focus of
this investigation was predominantly on cell cycle changes, and
bone remodeling changes were selected as a control. Data from the
scanned arrays were extracted using Genome Studios software (v 1.6;
Illumina, San Diego, CA, USA). Data processing and statistical
analysis were performed using MATLAB 2011a (MathWorks, Natick, MA,
USA), which was also used for multidimensional scaling analysis of
the detected probes. For gene analysis, the microarray data were
analyzed using the filtering criteria of adjusted P<0.05 and
>15% change. To reveal genes with significant changes in
expression, heatmap analysis was performed for visualization using
MATLAB.
Cell viability assay
A Cell Counting Kit-8 (Dojindo Molecular
Technologies, Inc., Kumamoto, Japan) was used to measure cell
viability following FGFR1 transfection. The MG63 cells
(103) were seeded into a 96-well plate and incubated for
48 h at 37°C in 5% CO2. At 48 h post-transfection, 10
µl CCK-8 solution was added to each well and the cells were
incubated for a further 4 h at 37°C, following which the absorbance
at 450 nm was measured using a microplate reader (Multiskan
Spectrum; Thermo Fisher Scientific). Each experiment was repeated
in triplicate, and observation of the cells was continued for 4
days to determine changes.
Statistical analysis
Data are representative of at least three
independent replicate experiments performed in triplicate, and are
expressed as the mean ± standard deviation. Student's t-test was
used to analyze the difference between two groups. P<0.05 was
considered to indicate a statistically significant difference. All
statistical analyses were performed with SPSS statistical software,
version 20.0 (IBM SPSS, Armonk, NY, USA).
Results
FGFR1 is expressed at high levels in
osteosarcoma tissues
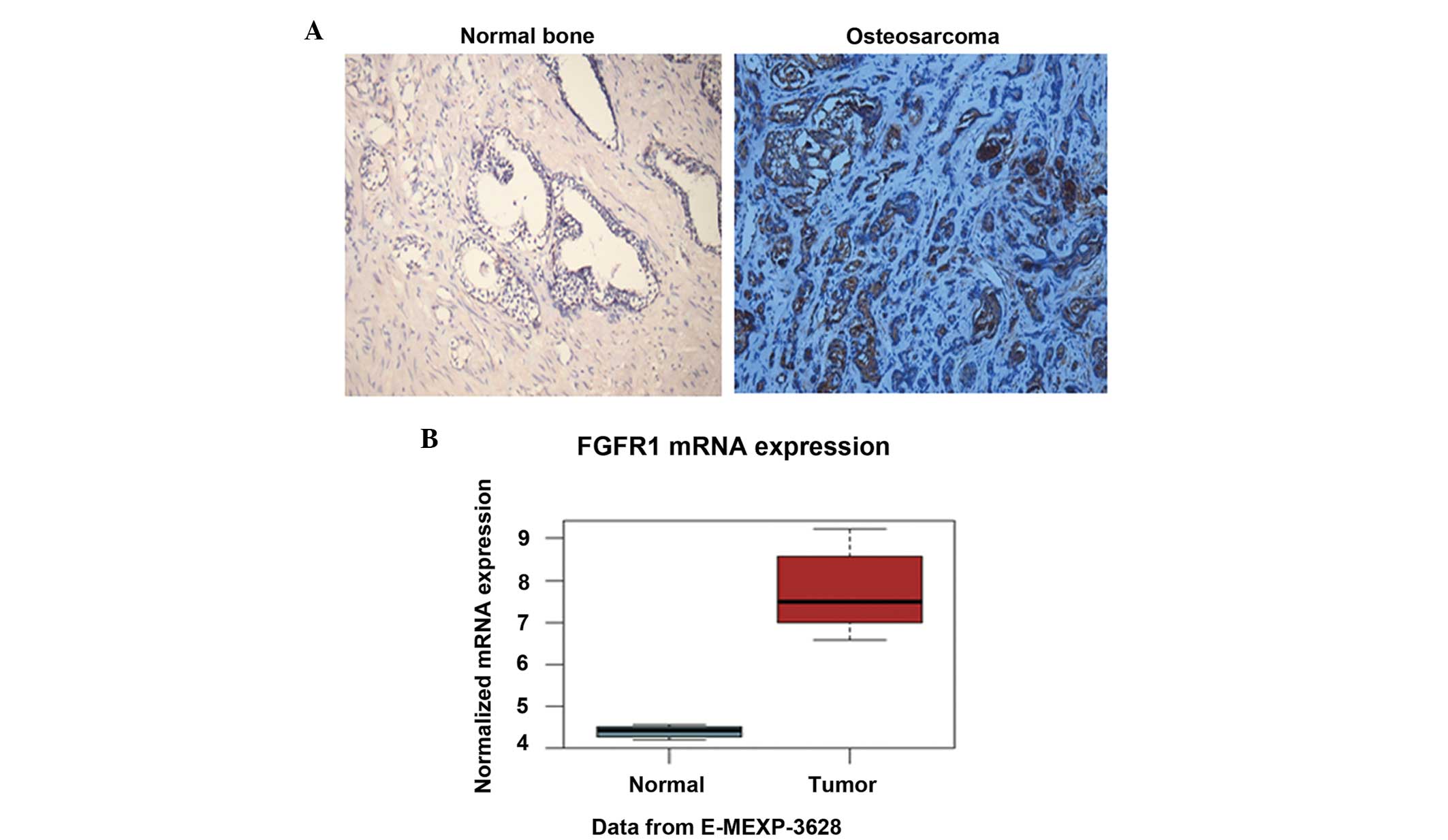
To determine the expression level of FGFR1 in
osteosarcoma tissues, the present study performed IHC analysis of
osteosarcoma tissues and paired normal bone tissues using the FGFR1
antibody. The results demonstrated that the protein level of FGFR1
in the osteosarcoma tissues was notably higher than that in the
paired normal bone tissues (Fig.
1A). The staining of FGFR1 was predominantly located in the
plasma (Fig. 1A). In addition, the
mRNA level of FGFR1 was determined in the two tissue groups.
Consistently, FGFR1 exhibited a relative mRNA level of ~4.5 in the
normal bone, whereas the relative mRNA level reached almost 7.5 in
the osteosarcoma tissues (Fig.
1B). The elevated protein and mRNA levels of FGFR1 in
osteosarcoma suggested that FGFR1 may be involved in
osteosarcomagenesis.
FGFR1 affects the cell cycle in
osteosarcoma
To investigate the possible pathways, which
contributed to the effects of FGFR in osteosarcoma, the Kyoto
Encyclopedia of Genes and Genomes (KEGG) database (http://www.kegg.jp/kegg/pathway.html)
was accessed (Fig. 2). Notably,
cell cycle was significantly affected in osteosarcoma (Fig. 2A(a); P=0.003). Bone remodeling is a
ubiquitous process in bone diseases, therefore, the bone remodeling
process was also assessed as a control. As expected, bone
remodeling was not significantly affected (Fig. 2A(b); P=0.369). As shown in Fig. 2C and D, the cell cycle was
significantly affected in osteosarcoma, with the upregulation of
multiple cell cycle-associated genes and the downregulation of
other genes downegulated. Furthermore, the present study performed
microarray analysis of osteosar-coma using MATLAB, and to identify
the genes and pathways that are involved in disease development,
heatmap analysis was performed. The heatmap analysis demonstrated
that multiple genes associated with the cell cycle were elevated,
whereas certain other genes were decreased in osteosarcoma
(Fig. 2B). Taken together, the
data from the KEGG and microarray analyses suggested that the cell
cycle is significantly affected by FGFR1 in osteosarcoma.
CDK1 is upregulated by FGFR1 in MG63
osteosarcoma cells
To further investigate the downstream molecules
regulated by FGFR1, the MG63 cells were transfected with the
Flag-FGFR1 expression plasmid. Transfection efficiency was
visualized using a fluorescence microscope (Fig. 3A). Subsequently, total RNA was
extracted and RT-qPCR analysis was performed to examine the
expression of cell cycle molecules, including CDK1 and CDK2.
Notably, the mRNA expression of CDK1 increased significantly in
response to FGFR1 overexpression, whereas no significant change was
observed in the mRNA level of CDK2 (Fig. 3B). Consistently, western blotting
also confirmed that the protein level of CDK1 was upregulated
following MG63 cell transfection with the pcDNA3-Flag-FGFR1 plasmid
(Fig. 3C). These data suggested
that FGFR1 targeted CDK1 to regulate the cell cycle in
osteosarcoma.
Overexpression of FGFR1 promotes MG63
cell proliferation
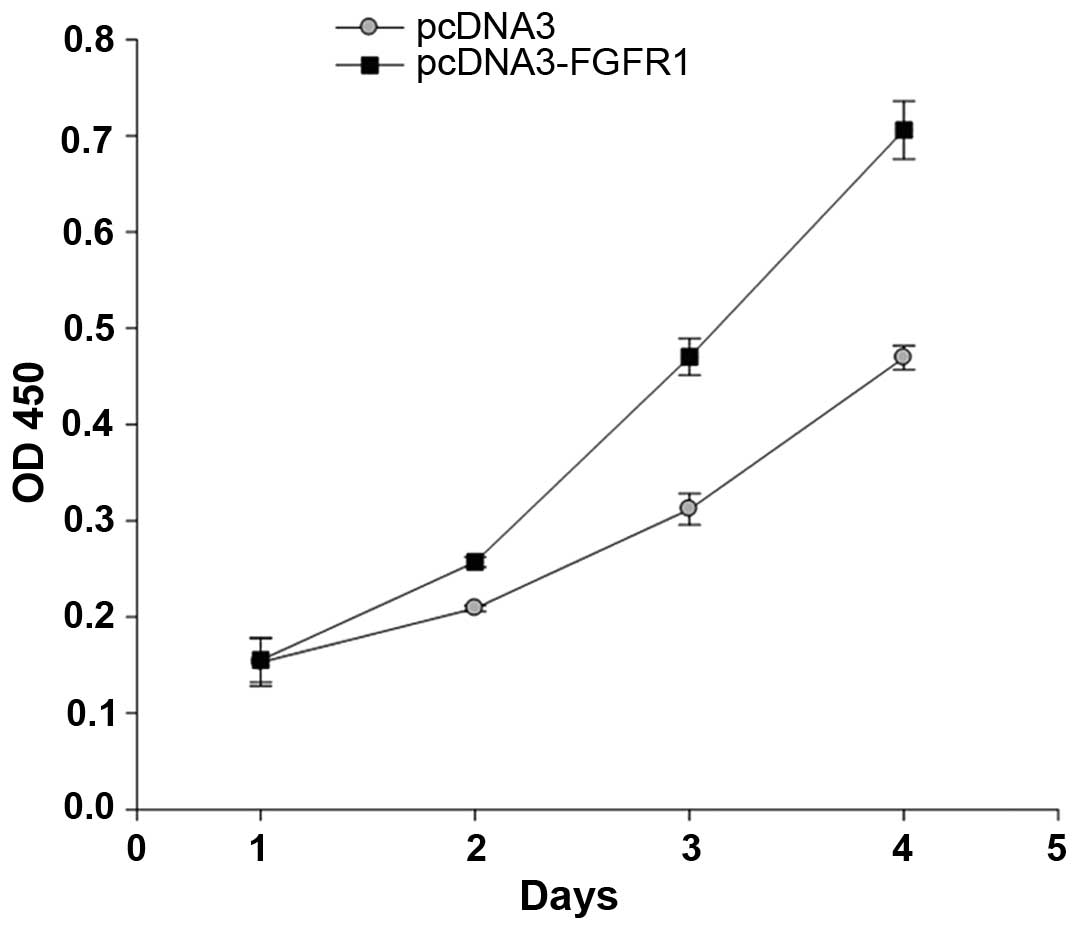
To investigate the role of FGFR1 in osteosarcoma
development, the MG63 cells were transfected with the
pcDNA3-Flag-FGFR1 plasmid and a cell viability assay was performed.
The absorbance at 450 nm, which represents cell numbers was
continuously determined for 5 days. The results demonstrated that
the number of cells in the FGFR1-overexpressed group were higher
than in the control group from day 2. Subsequently, the cell
numbers increased markedly, with the FGFR1-overexpressed MG63 cells
increasing ~7-fold by day 4. Notably, the number of
FGFR1-overexpressed MG63 cells increased by almost 1.7-fold,
compared with the control group on day 4 (Fig. 4). These data lead to the conclusion
that FGFR1 overexpression promoted MG63 cell growth.
Discussion
Although substantial progress has been achieved
through combination surgical resection and neoadjuvant chemotherapy
in the treatment of human osteosarcoma, the survival rate of
patients with osteosarcoma exhibiting localized diseases at
diagnosis has remained at ~70% since the mid-1980s, and the
long-term survival rate of patients with metastatic or recurrent
disease remains <20% (32,33).
Accordingly, increased understanding of the molecular mechanisms
underlying osteosarcomagenesis, and the identification of novel
molecular targets for osteosarcoma treatments, are urgently
required. The present study investigated the role of FGFR1 in the
development of human osteosarcoma, and examined the possible
mechanisms underlying the involvement of FGFR1 in osteosarcoma.
Consistent with the expression profile in other
types of cancer, the present study demonstrated that the expression
level of FGFR1 was elevated in osteosarcoma tissues (Fig. 1). Overexpression of FGFR1
significantly promoted MG63 cell proliferation (Fig. 4), indicating that a high expression
level of FGFR1 may be an indicator of osteosarcoma cell growth.
Furthermore, the possible pathways that contribute to the role of
FGFR1 in osteosarcoma were screened. KEGG database and heatmap
analysis were common strategies that were used for high-throughput
microarray analysis (34). With
the assistance of these two tools, the present study demonstrated
that cell cycle was significantly affected in the development of
FGFR1-mediated osteosarcoma (Fig.
2A). Multiple gene expression levels were elevated, and others
were decreased (Fig. 2B–D). Based
on these findings, the present study upregulated the expression of
FGFR in MG63 cells and, following verification of transfection
efficiency (Fig. 3A), detected the
expression of CDKs using RT-qPCR and western blot analysis. The
results revealed that the expression of CDK1, but not CDK2, was
significantly upregulated by FGFR1 in the MG63 cells (Fig. 3B and C). These findings indicated
that FGFR1 may affect cell cycle by regulating the expression of
CDK1.
CDKs are key regulators of the cell cycle and are
under tight control by external and internal signals (35). The targeting of CDKs is always
implicated in clinical anticancer drug developments (11,36).
The role of CDKs in neoplasia is also widely reported (11). In general, CDK1 and cyclin B1 are
associated with the G2/M phase; CDK2, cyclin E and cyclin A are
associated with the G1/S transition and S phase; and CDK4 and
cyclin D1 are associated with the G1 phase (37). In the present study, the
upregulation of FGFR1 caused a corresponding increase in the
expression of CDK1, without affecting the level of CDK2 (Fig. 3B and C). These data suggested that
FGFR1 may affect the G2/M phase. Of note, targeting CDK1, but not
CDK4/6 or CDK2, is selectively lethal to MYC-dependent human breast
cancer cells (38). Melatonin also
selectively targets CDK1, but not CDK2, to promote breast cancer
cell proliferation (37). These
reports confirm that the promoting effect of FGFR1 may be due to
the upregulation of CDK1, which is associated with the G2/M phase.
However, the detailed mechanisms of how FGFR1 regulates CDK1 and
affects the G2/M phase remain to be elucidated. As kinase activity
is critical for FGFR1 and CDK function, the present study
hypothesized that phosphorylation may be pivotal in FGFR1-mediated
cell cycle regulation. However, further investigation is
required.
The present study provided evidence that FGFR1
promoted osteosarcoma cell proliferation, and that CDK1 may be
critical in FGFR1-mediated osteosarcoma development. These findings
may improve current understanding of molecule mechanisms underlying
the development of osteosarcoma, and may provide novel insights
into the molecular targeted therapy for osteosarcoma. The
development of novel drugs targeting FGFR1 may be promising for
osteosarcoma treatment and prevention.
References
|
1
|
Longhi A, Errani C, De Paolis M, Mercuri M
and Bacci G: Primary bone osteosarcoma in the pediatric age: State
of the art. Cancer Treat Rev. 32:423–436. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kang K, Lee JH and Kim HG: Contralateral
referred pain in a patient with intramedullary spinal cord
metastasis from extraskeletal small cell osteosarcoma. J Spinal
Cord Med. 36:695–699. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chou AJ, Geller DS and Gorlick R: Therapy
for osteosarcoma: Where do we go from here? Paediatr Drugs.
10:315–327. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ahmad I, Iwata T and Leung HY: Mechanisms
of FGFR-mediated carcinogenesis. Biochim Biophys Acta.
1823.850–860. 2012.
|
|
5
|
Liang G, Liu Z, Wu J, Cai Y and Li X:
Anticancer molecules targeting fibroblast growth factor receptors.
Trends Pharmacol Sci. 33:531–541. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wesche J, Haglund K and Haugsten EM:
Fibroblast growth factors and their receptors in cancer. Biochem J.
437:199–213. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Johnson DE and Williams LT: Structural and
functional diversity in the FGF receptor multigene family. Adv
Cancer Res. 60:1–41. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chin K, DeVries S, Fridlyand J, Spellman
PT, Roydasgupta R, Kuo WL, Lapuk A, Neve RM, Qian Z, Ryder T, et
al: Genomic and transcriptional aberrations linked to breast cancer
pathophysiologies. Cancer Cell. 10:529–541. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gelsi-Boyer V, Orsetti B, Cervera N,
Finetti P, Sircoulomb F, Rougé C, Lasorsa L, Letessier A, Ginestier
C, Monville F, et al: Comprehensive profiling of 8p11-12
amplification in breast cancer. Mol Cancer Res. 3:655–667. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Jacquemier J, Adelaide J, Parc P,
Penault-Llorca F, Planche J, deLapeyriere O and Birnbaum D:
Expression of the FGFR1 gene in human breast-carcinoma cells. Int J
Cancer. 59:373–378. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
McDonald ER III and El-Deiry WS: Cell
cycle control as a basis for cancer drug development (Review). Int
J Oncol. 16:871–886. 2000.PubMed/NCBI
|
|
12
|
Roumiantsev S, Krause DS, Neumann CA,
Dimitri CA, Asiedu F, Cross NC and Van Etten RA: Distinct stem cell
myeloproliferative/T lymphoma syndromes induced by ZNF198-FGFR1 and
BCR-FGFR1 fusion genes from 8p11 trans-locations. Cancer Cell.
5:287–298. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Xiao S, Nalabolu SR, Aster JC, Ma J,
Abruzzo L, Jaffe ES, Stone R, Weissman SM, Hudson TJ and Fletcher
JA: FGFR1 is fused with a novel zinc-finger gene, ZNF198, in the
t(8;13) leukaemia/lymphoma syndrome. Nat Genet. 18:84–87. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yagasaki F, Wakao D, Yokoyama Y, Uchida Y,
Murohashi I, Kayano H, Taniwaki M, Matsuda A and Bessho M: Fusion
of ETV6 to fibroblast growth factor receptor 3 in peripheral T-cell
lymphoma with a t(4;12) (p16;p13) chromosomal translocation. Cancer
Res. 61:8371–8374. 2001.PubMed/NCBI
|
|
15
|
Clark JC, Tichelaar JW, Wert SE, Itoh N,
Perl AK, Stahlman MT and Whitsett J: FGF-10 disrupts lung
morphogenesis and causes pulmonary adenomas in vivo. Am J Physiol
Lung Cell Mol Physiol. 280:L705–L715. 2001.PubMed/NCBI
|
|
16
|
Marek L, Ware KE, Fritzsche A, Hercule P,
Helton WR, Smith JE, McDermott LA, Coldren CD, Nemenoff RA, Merrick
DT, et al: Fibroblast growth factor (FGF) and FGF receptor-mediated
autocrine signaling in non-small-cell lung cancer cells. Mol
Pharmacol. 75:196–207. 2009. View Article : Google Scholar :
|
|
17
|
van Rhijn BW, van Tilborg AA, Lurkin I,
Bonaventure J, de Vries A, Thiery JP, van der Kwast TH, Zwarthoff
EC and Radvanyi F: Novel fibroblast growth factor receptor 3
(FGFR3) mutations in bladder cancer previously identified in
non-lethal skeletal disorders. Eur J Hum Genet. 10:819–824. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Greenman C, Stephens P, Smith R, Dalgliesh
GL, Hunter C, Bignell G, Davies H, Teague J, Butler A, Stevens C,
et al: Patterns of somatic mutation in human cancer genomes.
Nature. 446:153–158. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Naski MC, Wang Q, Xu J and Ornitz DM:
Graded activation of fibroblast growth factor receptor 3 by
mutations causing achon-droplasia and thanatophoric dysplasia. Nat
Genet. 13:233–237. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Gru AA and Allred DC: FGFR1 amplification
and the progression of non-invasive to invasive breast cancer.
Breast Cancer Res. 14:1162012. View
Article : Google Scholar : PubMed/NCBI
|
|
21
|
Shiang CY, Qi Y, Wang B, Lazar V, Wang J,
Fraser Symmans W, Hortobagyi GN, Andre F and Pusztai L:
Amplification of fibroblast growth factor receptor-1 in breast
cancer and the effects of brivanib alaninate. Breast Cancer Res
Treat. 123:747–755. 2010. View Article : Google Scholar
|
|
22
|
Dutt A, Ramos AH, Hammerman PS, Mermel C,
Cho J, Sharifnia T, Chande A, Tanaka KE, Stransky N, Greulich H, et
al: Inhibitor-sensitive FGFR1 amplification in human non-small cell
lung cancer. PLoS One. 6:e203512011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Tran TN, Selinger CI, Kohonen-Corish MR,
McCaughan BC, Kennedy CW, O'Toole SA and Cooper WA: Fibroblast
growth factor receptor 1 (FGFR1) copy number is an independent
prognostic factor in non-small cell lung cancer. Lung Cancer.
81:462–467. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ren M, Hong M, Liu G, Wang H, Patel V,
Biddinger P, Silva J, Cowell J and Hao Z: Novel FGFR inhibitor
ponatinib suppresses the growth of non-small cell lung cancer cells
overexpressing FGFR1. Oncol Rep. 29:2181–2190. 2013.PubMed/NCBI
|
|
25
|
Acevedo VD, Gangula RD, Freeman KW, Li R,
Zhang Y, Wang F, Ayala GE, Peterson LE, Ittmann M and Spencer DM:
Inducible FGFR-1 activation leads to irreversible prostate
adenocarcinoma and an epithelial- to -mesenchymal transition.
Cancer Cell. 12:559–571. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lehnen NC, von Mässenhausen A, Kalthoff H,
Zhou H, Glowka T, Schütte U, Höller T, Riesner K, Boehm D,
Merkelbach-Bruse S, et al: Fibroblast growth factor receptor 1 gene
amplification in pancreatic ductal adenocarcinoma. Histopathology.
63:157–166. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Freier K, Schwaenen C, Sticht C,
Flechtenmacher C, Mühling J, Hofele C, Radlwimmer B, Lichter P and
Joos S: Recurrent FGFR1 amplification and high FGFR1 protein
expression in oral squamous cell carcinoma (OSCC). Oral Oncol.
43:60–66. 2007. View Article : Google Scholar
|
|
28
|
Simon R, Richter J, Wagner U, Fijan A,
Bruderer J, Schmid U, Ackermann D, Maurer R, Alund G, Knönagel H,
et al: High-throughput tissue microarray analysis of 3p25 (RAF1)
and 8p12 (FGFR1) copy number alterations in urinary bladder cancer.
Cancer Res. 61:4514–4519. 2001.PubMed/NCBI
|
|
29
|
Gorringe KL, Jacobs S, Thompson ER,
Sridhar A, Qiu W, Choong DY and Campbell IG: High-resolution single
nucleotide polymorphism array analysis of epithelial ovarian cancer
reveals numerous microdeletions and amplifications. Clin Cancer
Res. 13:4731–4739. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Missiaglia E, Selfe J, Hamdi M, Williamson
D, Schaaf G, Fang C, Koster J, Summersgill B, Messahel B, Versteeg
R, et al: Genomic imbalances in rhabdomyosarcoma cell lines affect
expression of genes frequently altered in primary tumors: An
approach to identify candidate genes involved in tumor development.
Genes Chromosomes Cancer. 48:455–467. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Guimarães JM, Guimarães IC, Duarte ME,
Vieira T, Vianna VF, Fernandes MB, Vieira AR and Casado PL:
Polymorphisms in BMP4 and FGFR1 genes are associated with fracture
non-union. J Orthop Res. 31:1971–1979. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Chou AJ, Merola PR, Wexler LH, Gorlick RG,
Vyas YM, Healey JH, LaQuaglia MP, Huvos AG and Meyers PA: Treatment
of osteosarcoma at first recurrence after contemporary therapy: The
memorial sloan-kettering cancer center experience. Cancer.
104:2214–2221. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
He H, Ni J and Huang J: Molecular
mechanisms of chemoresistance in osteosarcoma (Review). Oncol Lett.
7:1352–1362. 2014.PubMed/NCBI
|
|
34
|
Mougeot JL, Li Z, Price AE, Wright FA and
Brooks BR: Microarray analysis of peripheral blood lymphocytes form
ALS patients and the SAFE detection of the KEGG ALS pathway. BMC
Med Genomics. 25:74–80. 2011. View Article : Google Scholar
|
|
35
|
Morgan DO: Principles of CDK regulation.
Nature. 374:131–134. 1995. View
Article : Google Scholar : PubMed/NCBI
|
|
36
|
Benson C, Kaye S, Workman P, Garrett M,
Walton M and de Bono J: Clinical anticancer drug development:
Targeting the cyclin-dependent kinases. Br J Cancer. 92:7–12. 2005.
View Article : Google Scholar
|
|
37
|
Liu L, Xu Y and Reiter RJ: Melatonin
inhibits the proliferation of human osteosarcoma cell line MG-63.
Bone. 55:432–438. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kang J, Sergio CM, Sutherland RL and
Musgrove EA: Targeting cyclin-dependent kinase 1 (CDK1) but not
CDK4/6 or CDK2 is selectively lethal to MYC-dependent human breast
cancer cells. BMC Cancer. 14:322014. View Article : Google Scholar : PubMed/NCBI
|