Skeletal muscle makes up approximately 40% of the
body weight and is essential for locomotion (1). Skeletal muscle atrophy, predominantly
resulting from excessive protein degradation, occurs in various
conditions including starvation (2), aging (3), sepsis (4), cancer cachexia (5), severe burns (6,7) and
chronic kidney disease (8). Muscle
atrophy results in reductions in mobility of the patients and an
increased risk of mortality (9).
In patients with severe burns, skeletal muscle atrophy occurs as a
result of prolongation of time spent bed-bound and suppression of
wound healing (7). In general,
skeletal muscle atrophy predicts poor prognosis of patients.
The ubiquitin-proteasome pathway and cell apoptosis
are involved in regulating skeletal muscle atrophy (10–12).
The ubiquitin-proteasome pathway contributes to protein
degradation, as the targeted proteins for degradation are
substrates that can be identified and bound to ubiquitin.
Subsequently, poly-ubiquitinated substrates are targeted for
degradation by proteasomes (11).
Activation of the ubiquitin-proteasome pathway may serve an
important role in the mediation of skeletal muscle atrophy
(10,11). Previous studies have additionally
demonstrated that increased cell apoptosis is accompanied by
stress-induced skeletal muscle atrophy (12), and that apoptosis is also a
critical factor which leads to muscle atrophy (13,14).
The present review will focus upon the miRNAs
involved in the regulation of skeletal muscle atrophy and the
potential molecular mechanisms. Further studies are required in
order to elucidate the specific miRNAs implicated in stress-induced
skeletal muscle atrophy, which may lead to the development of novel
targets for clinical therapy.
Aberrant activation of protein degradation is the
key factor that leads to muscle atrophy, and the
ubiquitin-proteasome pathway serves a pivotal role in the mediation
of protein degradation (11). The
proteasome can identify the poly-ubiq-uitinated protein and trigger
the degradation procedure (23).
E3 ligase is the critical mediator of protein ubiquitination.
Muscle RING finger 1 (MuRF1) and muscle atrophy F-box (MAFbx) are
two muscle specific E3 ligases (24). During muscle atrophy, MuRF1 and
MAFbx are overexpressed in muscle, and inhibiting the function of
MuRF1 and MAFbx has been demonstrated to suppress muscle loss and
subsequently attenuate muscle atrophy (25,26).
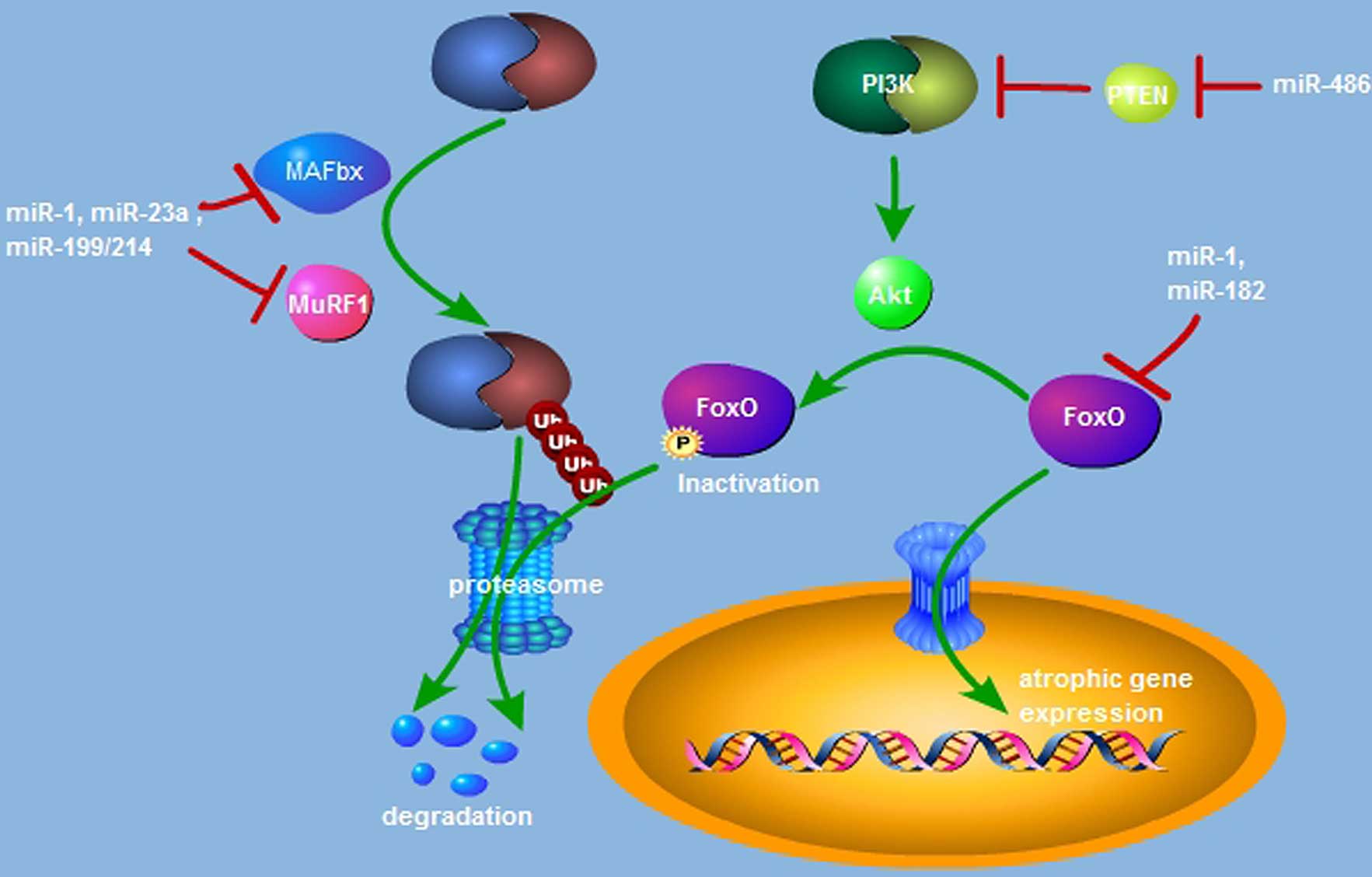
Previous studies (27–29) have additionally indicated that
miRNAs are implicated in the regulation of MuRF1 and MAFbx
expression (Fig. 1). miR-23a is
able to inhibit the translational activation of MuRF1 and MAFbx via
binding with their 3′-UTR, and miR-23a transgenic mice exert
resistance against glucocorticoid-induced muscle atrophy (27). In a dexamethasone (Dex)-induced
mouse model of atrophy, muscle-specific miR-1 expression is
upregulated. miR-1 has been previously reported to induce MuRF1 and
MAFbx expression via the HSP70/protein kinase B(Akt)/forkhead box
(Fox) O3 signaling pathway and is responsible for Dex-induced
muscle atrophy (28). The
miR-199/214 cluster is also involved in regulating the
ubiquitin-proteasome pathway (29). Taken together, miRNA-dependent
activation of the ubiquitin-proteasome pathway is responsible for
the promotion of muscle atrophy by directly or indirectly targeting
the muscle specific E3 ligases of MuRF1 and MAFbx.
In addition to enhancing muscle proteolysis,
aberrant myogenesis is also a critical factor during muscle
atrophy. Myogenesis is impaired in models of mice with cancer
(30), and pigs with chronic
obstructive pulmonary disease (31). Inactivation of myogenesis is also
observed in diseases such as Duchenne muscular dystrophy and spinal
and bulbar muscular atrophy (32,33).
Muscle satellite cells are stem cells with self-renewal and
differentiation potency, and when muscle disruption occurs,
proliferative satellite cells could differentiate into myotubes and
contribute to muscle regeneration (34). Defects in post-natal myogenesis and
muscle regeneration result in muscle atrophy, and miRNAs are
implicated in the regulation of myogenesis and muscle atrophy
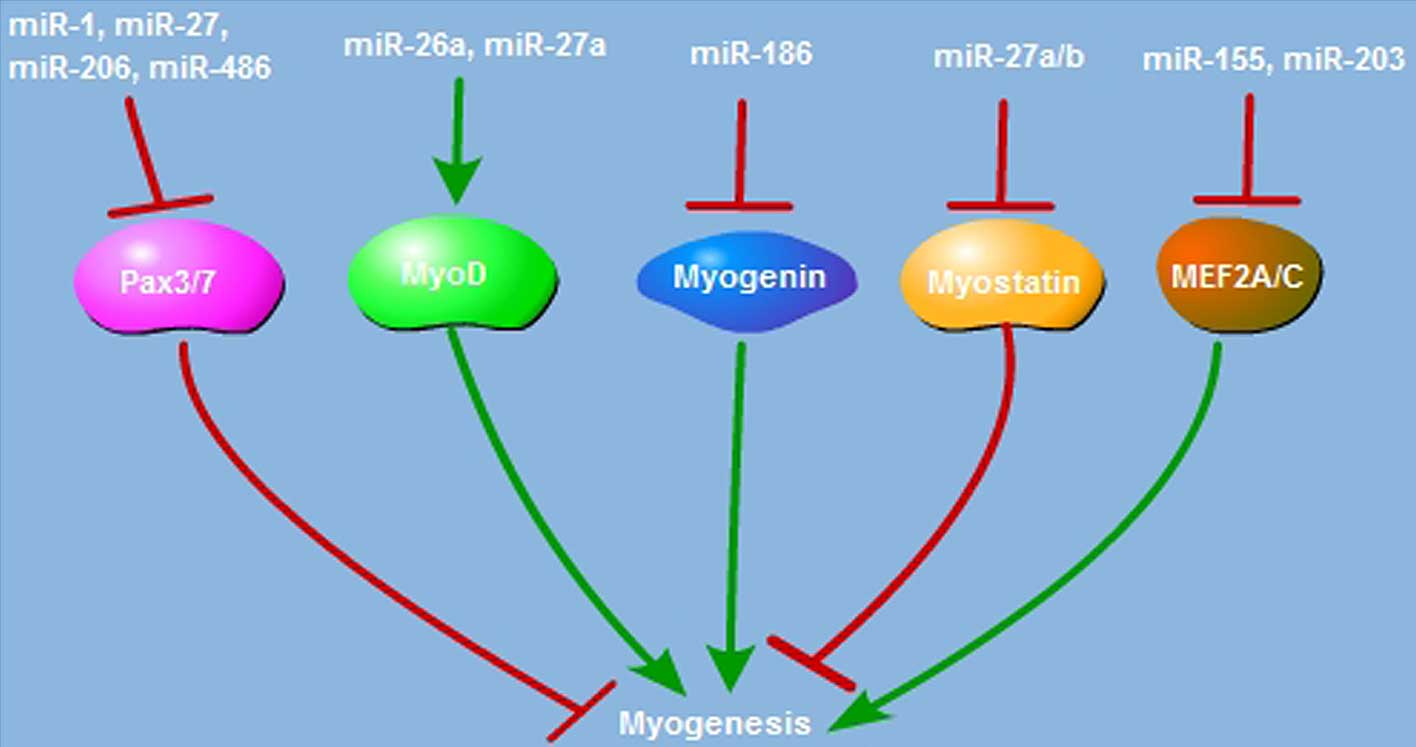
(35,36). As presented in Fig. 2, paired-box transcription factor
(Pax) is essential for satellite cell proliferation and
differentiation. miR-1, miR-206 and miR-486 restrict satellite cell
proliferation and promote its differentiation through suppression
of Pax7 expression (37–39). Pax3 is also the critical factor
required to trigger satellite cell proliferation; suppressing
miR-27b, miR-1 and miR-206 expression suppresses satellite cell
differentiation via enhancement of Pax3 activation (40,41).
The myogenic regulatory factor (MRF) family, which includes MyoD,
Myf5, myogenin and Myf6, has the pivotal role in myogenic
differentiation. MyoD is expressed in activated satellite cells,
and miR-27a overexpression elevates the MyoD protein level and
enhances myoblast differentiation (42). In C2C12 myoblast cells, miR-26a
upregulates MyoD expression and promotes the myogenic process
(43). Subsequent to injury,
miR-26a is induced during muscle regeneration, and blocking miR-26a
expression enhances Smad-dependent muscle differentiation (44). miR-186 suppresses C2C12 myoblast
cell myogenic differentiation via targeting myogenin (45). In addition to the MRF family,
miRNAs also regulate myogenesis through targeting a variety of
proteins. Myostatin is the negative mediator of myogenesis; miR-27a
and miR-27b promote satellite cell proliferation and post-natal
myogenesis by suppressing myostatin expression (46–48).
miR-125b, miR-133 and miR-199a-3p are involved in the regulation of
the insulin-like growth factor/insulin-like growth factor receptor
signaling pathway and inhibit cell differentiation and muscle
regeneration (49–51). miR-203 functions as the suppressor
of myoblast differentiation by repressing c-Jun and myocyte
enhancer factor 2C (MEF2C) expression (52). miR-155 inhibits MEF2A expression
and suppresses the myogenic process (53). miR-29 is a pro-myogenic factor,
which acts through downregulation of Akt3 or RING1 and YY1-binding
protein (54,55). Thus, miRNAs have critical roles in
regulating satellite cell proliferation, myogenic differentiation
and muscle regeneration.
Cell apoptosis is programmed cell death and is a
promoting factor of muscle atrophy (14,56).
Studies using a mouse model have demonstrated that cell apoptosis
is involved in the progression of heart failure, severe burns and
age-associated muscle atrophy (57–60).
The mitochondria and caspase-mediated apoptotic pathways are some
of the mechanisms of burn, age or stress-induced muscle atrophy
(12,57,61,62).
miRNA is an important mediator of myoblast cell apoptosis (63). In skeletal muscle, pre-conditional
activation of interleukin (IL)-11/signal transducer and activator
of transcription (STAT)3 pathway protects human skeletal myoblasts
from oxidant-induced apoptosis (64), and miR-21 is a key regulator of
extracellular signal-related kinase 1/2-STAT3 signaling downstream
of IL-11 and inhibits the apoptosis of skeletal myoblasts (65). Skeletal muscle loss in cancer
cachexia is partially associated with cell apoptosis, and a
previous study indicated that miR-21 in microvesicles of cancer
cachexia triggers muscle cell apoptosis via enhancement of c-Jun
N-terminal kinase activation (66). In acute muscle injury, myogenic
progenitor cell apoptosis is triggered by miR-351 knockdown
(21). MyoD is a critical factor
in the regulation muscle differentiation; MyoD knockout in
myoblasts decelerates miR-1 and miR-206 expression and results in
resistance to apoptosis (67).
Forced MyoD expression in MyoD knockout myoblasts enhances the
expression of miR-1 and miR-206 and triggers cell apoptosis via
Pax3 downregulation (67). Thus,
it is suggested that miRNA is a critical mediator in regulating
myoblast apoptosis and implicated in muscle atrophic process.
Aberrant muscle protein degradation, impairment of
myogenesis, and promotion of muscle cell apoptosis are all
important factors that contribute to muscle atrophy. miRNAs are
critical mediators of protein degradation and myogenesis through
regulation of the ubiquitin-proteasome and PI3K/Akt/FoxO signaling
pathways and other myogenic regulatory factors. Thus, miRNAs may be
potential and effective therapeutic targets for muscle atrophy.
The authors would like to thank Dr Hongli Wu for her
critical reading of this review. The current study was supported by
the National Science Foundation of China (grant nos.
NSFC81120108014, NSFC81471873, NSFC81501694, NSFC81571894 and
NSFC81171807), the Beijing Natural Science Foundation (grant no.
7144250) and the Nursery Fund of People's Liberation Army General
Hospital (grant nos. 14KMM22 and 13KMM31).
|
1
|
Hitachi K and Tsuchida K: Role of
microRNAs in skeletal muscle hypertrophy. Front Physiol. 4:4082014.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Paul PK, Bhatnagar S, Mishra V, Srivastava
S, Darnay BG, Choi Y and Kumar A: The E3 Kubiquitin ligase TRAF6
intercedes in starvation-induced skeletal muscle atrophy through
multiple mechanisms. Mol Cell Biol. 32:1248–1259. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
McGregor RA, Poppitt SD and Cameron-Smith
D: Role of microRNAs in the age-related changes in skeletal muscle
and diet or exercise interventions to promote healthy aging in
humans. Ageing Res Rev. 17:25–33. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Nystrom G, Pruznak A, Huber D, Frost RA
and Lang CH: Local insulin-like growth factor I prevents
sepsis-induced muscle atrophy. Metabolism. 58:787–797. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wang H, Lai YJ, Chan YL, Li TL and Wu CJ:
Epigallocatechin-3-gallate effectively attenuates skeletal muscle
atrophy caused by cancer cachexia. Cancer Lett. 305:40–49. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bertsch S, Lang CH and Vary TC: Inhibition
of glycogen synthase kinase 3[beta] activity with lithium in vitro
attenuates sepsis-induced changes in muscle protein turnover.
Shock. 35:266–274. 2011. View Article : Google Scholar
|
|
7
|
Hart DW, Wolf SE, Chinkes DL, Gore DC,
Mlcak RP, Beauford RB, Obeng MK, Lal S, Gold WF, Wolfe RR and
Herndon DN: Determinants of skeletal muscle catabolism after severe
burn. Ann Surg. 232:455–465. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Xu J, Li R, Workeneh B, Dong Y, Wang X and
Hu Z: Transcription factor FoxO1, the dominant mediator of muscle
wasting in chronic kidney disease, is inhibited by microRNA-486.
Kidney Int. 82:401–411. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Metter EJ, Talbot LA, Schrager M and
Conwit R: Skeletal muscle strength as a predictor of all-cause
mortality in healthy men. J Gerontol A Biol Sci Med Sci.
57:B359–B365. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chai J, Wu Y and Sheng ZZ: Role of
ubiquitin-proteasome pathway in skeletal muscle wasting in rats
with endotoxemia. Crit Care Med. 31:1802–1807. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Attaix D, Combaret L, Bechet D and
Taillandier D: Role of the ubiquitin-proteasome pathway in muscle
atrophy in cachexia. Curr Opin Support Palliat Care. 2:262–266.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sishi B, Loos B, Ellis B, Smith W, du Toit
EF and Engelbrecht AM: Diet-induced obesity alters signalling
pathways and induces atrophy and apoptosis in skeletal muscle in a
prediabetic rat model. Exp Physiol. 96:179–193. 2011. View Article : Google Scholar
|
|
13
|
Engelbrecht AM, Smith C, Neethling I,
Thomas M, Ellis B, Mattheyse M and Myburgh KH: Daily brief
restraint stress alters signaling pathways and induces atrophy and
apoptosis in rat skeletal muscle. Stress. 13:132–141. 2010.
View Article : Google Scholar
|
|
14
|
Dupont-Versteegden EE: Apoptosis in
skeletal muscle and its relevance to atrophy. World J
Gastroenterol. 12:7463–7466. 2006.PubMed/NCBI
|
|
15
|
Lee RC, Feinbaum RL and Ambros V: The C.
elegans heterochronic gene lin-4 encodes small RNAs with antisense
complementarity to lin-14. Cell. 75:843–854. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sayed D and Abdellatif M: MicroRNAs in
development and disease. Physiol Rev. 91:827–887. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Didiano D and Hobert O: Molecular
architecture of a miRNA-regulated 3′ UTR. RNA. 14:1297–1317. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Travaglini L, Vian L, Billi M, Grignani F
and Nervi C: Epigenetic reprogramming of breast cancer cells by
valproic acid occurs regardless of estrogen receptor status. Int J
Biochem Cell Biol. 41:225–234. 2009. View Article : Google Scholar
|
|
20
|
Taulli R, Bersani F, Foglizzo V, Linari A,
Vigna E, Ladanyi M, Tuschl T and Ponzetto C: The muscle-specific
microRNA miR-206 blocks human rhabdomyosarcoma growth in
xeno-transplanted mice by promoting myogenic differentiation. J
Clin Invest. 119:2366–2378. 2009.PubMed/NCBI
|
|
21
|
Chen Y, Melton DW, Gelfond JA, McManus LM
and Shireman PK: MiR-351 transiently increases during muscle
regeneration and promotes progenitor cell proliferation and
survival upon differentiation. Physiol Genomics. 44:1042–1051.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Motohashi N, Alexander MS,
Shimizu-Motohashi Y, Myers JA, Kawahara G and Kunkel LM: Regulation
of IRS1/Akt insulin signaling by microRNA-128a during myogenesis. J
Cell Sci. 126:2678–2691. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hartmann-Petersen R and Gordon C: Proteins
interacting with the 26S proteasome. Cell Mol Life Sci.
61:1589–1595. 2004.PubMed/NCBI
|
|
24
|
Bodine SC, Latres E, Baumhueter S, Lai VK,
Nunez L, Clarke BA, Poueymirou WT, Panaro FJ, Na E, Dharmarajan K,
et al: Identification of ubiquitin ligases required for skeletal
muscle atrophy. Science. 294:1704–1708. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Eddins MJ, Marblestone JG, Suresh Kumar
KG, Leach CA, Sterner DE, Mattern MR and Nicholson B: Targeting the
ubiquitin E3 ligase MuRF1 to inhibit muscle atrophy. Cell Biochem
Biophys. 60:113–118. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Clavel S, Coldefy AS, Kurkdjian E, Salles
J, Margaritis I and Derijard B: Atrophy-related ubiquitin ligases,
atrogin-1 and MuRF1 are up-regulated in aged rat Tibialis Anterior
muscle. Mech Ageing Dev. 127:794–801. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wada S, Kato Y, Okutsu M, Miyaki S, Suzuki
K, Yan Z, Schiaffino S, Asahara H, Ushida T and Akimoto T:
Translational suppression of atrophic regulators by microRNA-23a
integrates resistance to skeletal muscle atrophy. J Biol Chem.
286:38456–38465. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kukreti H, Amuthavalli K, Harikumar A,
Sathiyamoorthy S, Feng PZ, Anantharaj R, Tan SL, Lokireddy S,
Bonala S, Sriram S, et al: Muscle-specific microRNA1 (miR1) targets
heat shock protein 70 (HSP70) during dexamethasone-mediated
atrophy. J Biol Chem. 288:6663–6678. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Baumgarten A, Bang C, Tschirner A,
Engelmann A, Adams V, von Haehling S, Doehner W, Pregla R, Anker
MS, Blecharz K, et al: TWIST1 regulates the activity of ubiquitin
proteasome system via the miR-199/214 cluster in human end-stage
dilated cardiomyopathy. Int J Cardiol. 168:1447–1452. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Penna F, Costamagna D, Fanzani A, Bonelli
G, Baccino FM and Costelli P: Muscle wasting and impaired
myogenesis in tumor bearing mice are prevented by ERK inhibition.
PLoS One. 5:e136042010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Verhees KJ, Pansters NA, Baarsma HA,
Remels AH, Haegens A, de Theije CC, Schols AM, Gosens R and Langen
RC: Pharmacological inhibition of GSK-3 in a guinea pig model of
LPS-induced pulmonary inflammation: II. Effects on skeletal muscle
atrophy. Respir Res. 14:1172013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Shi H, Verma M, Zhang L, Dong C, Flavell
RA and Bennett AM: Improved regenerative myogenesis and muscular
dystrophy in mice lacking Mkp5. J Clin Invest. 123:2064–2077. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Malena A, Pennuto M, Tezze C, Querin G,
D'Ascenzo C, Silani V, Cenacchi G, Scaramozza A, Romito S, Morandi
L, et al: Androgen-dependent impairment of myogenesis in spinal and
bulbar muscular atrophy. Acta Neuropathol. 126:109–121. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Sacco A, Doyonnas R, Kraft P, Vitorovic S
and Blau HM: Self-renewal and expansion of single transplanted
muscle stem cells. Nature. 456:502–506. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Dachs E, Hereu M, Piedrafita L, Casanovas
A, Calderó J and Esquerda JE: Defective neuromuscular junction
organization and postnatal myogenesis in mice with severe spinal
muscular atrophy. J Neuropathol Exp Neurol. 70:444–461. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wang XH: MicroRNA in myogenesis and muscle
atrophy. Curr Opin Clin Nutr Metab Care. 16:258–266. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Chen JF, Tao Y, Li J, Deng Z, Yan Z, Xiao
X and Wang DZ: microRNA-1 and microRNA-206 regulate skeletal muscle
satellite cell proliferation and differentiation by repressing
Pax7. J Cell Biol. 190:867–879. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Dey BK, Gagan J and Dutta A: miR-206 and
-486 induce myoblast differentiation by downregulating Pax7. Mol
Cell Biol. 31:203–214. 2011. View Article : Google Scholar :
|
|
39
|
Liu N, Williams AH, Maxeiner JM,
Bezprozvannaya S, Shelton JM, Richardson JA, Bassel-Duby R and
Olson EN: microRNA-206 promotes skeletal muscle regeneration and
delays progression of Duchenne muscular dystrophy in mice. J Clin
Invest. 122:2054–2065. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
40
|
Goljanek-Whysall K, Sweetman D, Abu-Elmagd
M, Chapnik E, Dalmay T, Hornstein E and Münsterberg A: MicroRNA
regulation of the paired-box transcription factor Pax3 confers
robustness to developmental timing of myogenesis. Proc Natl Acad
Sci USA. 108:11936–11941. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Crist CG, Montarras D, Pallafacchina G,
Cumano A, Conway SJ and Buckingham M: Muscle stem cell behavior is
modified by microRNA-27 regulation of Pax3 expression. Proc Natl
Acad Sci USA. 106:13383–13387. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Chen X, Huang Z, Chen D, Yang T and Liu G:
Role of microRNA-27a in myoblast differentiation. Cell Biol Int.
38:266–271. 2014. View Article : Google Scholar
|
|
43
|
Wong CF and Tellam RL: MicroRNA-26a
targets the histone methyltransferase Enhancer of Zeste homolog 2
during myogenesis. J Biol Chem. 283:9836–9843. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Dey BK, Gagan J, Yan Z and Dutta A:
miR-26a is required for skeletal muscle differentiation and
regeneration in mice. Genes Dev. 26:2180–2191. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Antoniou A, Mastroyiannopoulos NP, Uney JB
and Phylactou LA: miR-186 inhibits muscle cell differentiation
through myogenin regulation. J Biol Chem. 289:3923–3935. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Huang Z, Chen X, Yu B, He J and Chen D:
MicroRNA-27a promotes myoblast proliferation by targeting
myostatin. Biochem Biophys Res Commun. 423:265–269. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
McFarlane C, Vajjala A, Arigela H,
Lokireddy S, Ge X, Bonala S, Manickam R, Kambadur R and Sharma M:
Negative auto-regulation of myostatin expression is mediated by
Smad3 and microRNA-27. PLoS One. 9:e876872014. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Yang T, Chen XL, Huang ZQ, Wen WX, Xu M,
Chen DW, Yu B, He J, Luo JQ, Yu J, et al: MicroRNA-27a promotes
porcine myoblast proliferation by downregulating myostatin
expression. Animal. 8:1867–1872. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Ge Y, Sun Y and Chen J: IGF-II is
regulated by microRNA-125b in skeletal myogenesis. J Cell Biol.
192:69–81. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Huang MB, Xu H, Xie SJ, Zhou H and Qu LH:
Insulin-like growth factor-1 receptor is regulated by microRNA-133
during skeletal myogenesis. PLoS One. 6:e291732011. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Jia L, Li YF, Wu GF, Song ZY, Lu HZ, Song
CC, Zhang QL, Zhu JY, Yang GS and Shi XE: MiRNA-199a-3p regulates
C2C12 myoblast differentiation through IGF-1/AKT/mTOR signal
pathway. Int J Mol Sci. 15:296–308. 2013. View Article : Google Scholar
|
|
52
|
Luo W, Wu H, Ye Y, Li Z, Hao S, Kong L,
Zheng X, Lin S, Nie Q and Zhang X: The transient expression of
miR-203 and its inhibiting effects on skeletal muscle cell
proliferation and differentiation. Cell Death Dis. 5:e13472014.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Seok HY, Tatsuguchi M, Callis TE, He A, Pu
WT and Wang DZ: miR-155 inhibits expression of the MEF2A protein to
repress skeletal muscle differentiation. J Biol Chem.
286:35339–35346. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Wei W, He HB, Zhang WY, Zhang HX, Bai JB,
Liu HZ, Cao JH, Chang KC, Li XY and Zhao SH: miR-29 targets Akt3 to
reduce proliferation and facilitate differentiation of myoblasts in
skeletal muscle development. Cell Death Dis. 4:e6682013. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Zhou L, Wang L, Lu L, Jiang P, Sun H and
Wang H: A novel target of microRNA-29, Ring1 and YY1-binding
protein (Rybp), negatively regulates skeletal myogenesis. J Biol
Chem. 287:25255–25265. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Dupont-Versteegden EE: Apoptosis in muscle
atrophy: Relevance to sarcopenia. Exp Gerontol. 40:473–481. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Dirks AJ and Leeuwenburgh C: The role of
apoptosis in age-related skeletal muscle atrophy. Sports Med.
35:473–483. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Lee HY, Kaneki M, Andreas J, Tompkins RG
and Martyn JA: Novel mitochondria-targeted antioxidant peptide
ameliorates burn-induced apoptosis and endoplasmic reticulum stress
in the skeletal muscle of mice. Shock. 36:580–585. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Fanzani A, Conraads VM, Penna F and
Martinet W: Molecular and cellular mechanisms of skeletal muscle
atrophy: An update. J Cachexia Sarcopenia Muscle. 3:163–179. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Libera LD, Zennaro R, Sandri M, Ambrosio
GB and Vescovo G: Apoptosis and atrophy in rat slow skeletal
muscles in chronic heart failure. Am J Physiol. 277:C982–C986.
1999.PubMed/NCBI
|
|
61
|
Yasuhara S, Perez ME, Kanakubo E, Yasuhara
Y, Shin YS, Kaneki M, Fujita T and Martyn JA: Skeletal muscle
apoptosis after burns is associated with activation of proapoptotic
signals. Am J Physiol Endocrinol Metab. 279:E1114–E1121.
2000.PubMed/NCBI
|
|
62
|
Marzetti E, Lawler JM, Hiona A, Manini T,
Seo AY and Leeuwenburgh C: Modulation of age-induced apoptotic
signaling and cellular remodeling by exercise and calorie
restriction in skeletal muscle. Free Radic Biol Med. 44:160–168.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Callis TE, Chen JF and Wang DZ: MicroRNAs
in skeletal and cardiac muscle development. DNA Cell Biol.
26:219–225. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Idris NM, Ashraf M, Ahmed RP, Shujia J and
Haider KH: Activation of IL-11/STAT3 pathway in preconditioned
human skeletal myoblasts blocks apoptotic cascade under oxidant
stress. Regen Med. 7:47–57. 2012. View Article : Google Scholar
|
|
65
|
Haider KH, Idris NM, Kim HW, Ahmed RP,
Shujia J and Ashraf M: MicroRNA-21 is a key determinant in
IL-11/Stat3 anti-apoptotic signalling pathway in preconditioning of
skeletal myoblasts. Cardiovasc Res. 88:168–178. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
He WA, Calore F, Londhe P, Canella A,
Guttridge DC and Croce CM: Microvesicles containing miRNAs promote
muscle cell death in cancer cachexia via TLR7. Proc Natl Acad Sci
USA. 111:4525–4529. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Hirai H, Verma M, Watanabe S, Tastad C,
Asakura Y and Asakura A: MyoD regulates apoptosis of myoblasts
through microRNA-mediated down-regulation of Pax3. J Cell Biol.
191:347–365. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Stitt TN, Drujan D, Clarke BA, Panaro F,
Timofeyva Y, Kline WO, Gonzalez M, Yancopoulos GD and Glass DJ: The
IGF-1/PI3K/Akt pathway prevents expression of muscle
atrophy-induced ubiquitin ligases by inhibiting FOXO transcription
factors. Mol Cell. 14:395–403. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Sugita H, Kaneki M, Sugita M, Yasukawa T,
Yasuhara S and Martyn JA: Burn injury impairs insulin-stimulated
Akt/PKB activation in skeletal muscle. Am J Physiol Endocrinol
Metab. 288:E585–E591. 2005. View Article : Google Scholar
|
|
70
|
Du K, Yu Y, Zhang D, Luo W, Huang H, Chen
J, Gao J and Huang C: NFkappaB1 (p50) suppresses SOD2 expression by
inhibiting FoxO3a transactivation in a miR190/PHLPP1/Akt-dependent
axis. Mol Biol Cell. 24:3577–3583. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Sandri M, Sandri C, Gilbert A, Skurk C,
Calabria E, Picard A, Walsh K, Schiaffino S, Lecker SH and Goldberg
AL: Foxo transcription factors induce the atrophy-related ubiquitin
ligase atrogin-1 and cause skeletal muscle atrophy. Cell.
117:399–412. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Sheriff S, Kadeer N, Joshi R, Friend LA,
James JH and Balasubramaniam A: Des-acyl ghrelin exhibits
pro-anabolic and anti-catabolic effects on C2C12 myotubes exposed
to cytokines and reduces burn-induced muscle proteolysis in rats.
Mol Cell Endocrinol. 351:286–295. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Alexander MS, Casar JC, Motohashi N, Myers
JA, Eisenberg I, Gonzalez RT, Estrella EA, Kang PB, Kawahara G and
Kunkel LM: Regulation of DMD pathology by an ankyrin-encoded miRNA.
Skelet Muscle. 1:272011. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Chen D, Goswami CP, Burnett RM, Anjanappa
M, Bhat-Nakshatri P, Muller W and Nakshatri H: Cancer affects
microRNA expression, release and function in cardiac and skeletal
muscle. Cancer Res. 74:4270–4281. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Hitachi K, Nakatani M and Tsuchida K:
Myostatin signaling regulates Akt activity via the regulation of
miR-486 expression. Int J Biochem Cell Biol. 47:93–103. 2014.
View Article : Google Scholar
|
|
76
|
Hudson MB, Rahnert JA, Zheng B,
Woodworth-Hobbs ME, Franch HA and Price SR: miR-182 attenuates
atrophy-related gene expression by targeting FoxO3 in skeletal
muscle. Am J Physiol Cell Physiol. 307:C314–C319. 2014. View Article : Google Scholar : PubMed/NCBI
|