Introduction
Osteoporosis occurs due to an imbalanced homeostasis
between bone resorption osteoclasts and bone formation osteoblasts
and represents a global health problem. The low bone mineral
density observed in post-menopausal women is mainly due to enhanced
osteoclast formation and/or function. Previous studies have
demonstrated that numerous key cytokines have important roles in
osteoclastogenesis and osteoporosis, including macrophage
colony-stimulating factor (M-CSF), receptor activator of nuclear
factor-κB ligand (RANKL), interleukin 1 and tumor necrosis factor α
(1–3). Consequently, several of these key
cytokines have been developed as therapeutic targets for the
treatment of osteoporosis. For instance, denosumab, which targets
RANKL, has been proved to effectively prevent osteoporosis in
post-menopausal women (4).
Apart from cytokines, microRNAs (miRNAs/miRs) have
essential roles in regulating cell biology and have been proposed
as potential biomarkers or therapeutic targets for several diseases
(5–7). miRNAs are non-coding small RNAs
containing ~22 nucleotides. By suppressing translation of the
targeted mRNAs, they are estimated to control more than one third
of all human genes (8). Previous
studies have demonstrated that miRNAs have critical roles in
regulating osteoclastogenesis and are associated with numerous
osteolytic diseases (9–14). Considering the possible adverse
effects of the synthetic drugs currently used for the treatment of
these diseases, miRNAs have received increased attention as
possible alternative therapeutic targets for the prevention and
treatment of osteolytic diseases such as osteoporosis (13,14).
Therefore, the present study aimed to assess differentially
expressed miRNAs during osteoclastogenesis in vitro and in
ovariectomy (OVX)-induced osteoporosis in vivo. More
specifically, the present study focused on the downregulated
miRNAs, which were associated with the progression of osteoporosis
in vivo.
Materials and methods
Media and reagents
The α-modified Eagle's medium (α-MEM), foetal bovine
serum (FBS) and penicillin/streptomycin were obtained from Gibco
(Thermo Fisher Scientific, Inc., Waltham, MA, USA). The Cell
Counting kit-8 (CCK-8) was purchased from Dojindo Molecular
Technology (Kumamoto, Japan). Mouse RANKL and recombinant soluble
mouse M-CSF were obtained from R&D Systems (Minneapolis, MN,
USA). The tartrate-resistant acid phosphatase (TRAP) staining kit,
TRIzol, chloral hydrate and ketamine were purchased from
Sigma-Aldrich (St Louis, MO, USA). All other reagents were from
Sigma-Aldrich, unless stated otherwise. The present study was
approved by the ethics committee of Sir Run Run Shaw Hospital
(Hangzhou, China).
Bone marrow cell culture
Bone marrow macrophages (BMMs) were separated from
the femurs and tibiae of two six-week-old female C57BL/6 mice
(Shanghai SLAC Laboratory Animal Co., Ltd., Shanghai, China) as
described previously (15). The
isolated BMMs were then cultured in α-MEM containing 10% FBS and 30
ng/ml M-CSF for proliferation. Cells were harvested when reaching
90% confluence and seeded into 96-well plates at a density of
8×103 cells/well or into six-well plates at a density of
8×105 cells/well in triplicate. After incubation for 24
h, cells were treated with RANKL (50 ng/ml) for various durations
(0, 1, 3, 5 or 7 days). Cells in 96-well plates were then fixed and
stained for TRAP activity following the kit manufacturer's
instructions. Cells in the six-well plates were dissolved in TRIzol
for extraction of total RNA (including miRNA).
Animal model
An OVX-induced osteoporosis model was used to
establish various degrees of osteoporosis in vivo. All
experimental procedures were approved by the Animal Care and
Experiment Committee of Zhejiang University School of Medicine
(Huangzhou, China), and the study was performed according to the
guidelines for Ethical Conduct from the Care and Use of Nonhuman
Animals in Research by the American Psychological Association.
Healthy eight-week-old C57BL/6J mice (n=24) were subjected to sham
operation (n=8) or ovariectomy (OVX) (n=16) under anesthesia using
8% chloral hydrate. The mice were housed in three cages in an
animal facility with conditions of 22–24°C, 12-h light/dark cycle,
50–60% humidity with access to food and water ad libitum.
Eight sham-operated mice and eight OVX mice were sacrificed by
euthanasia with 0.8 ml/100 g body weight ketamine delivered by
intraperitoneal injection at 1 month following surgery (sham and
OVX 1). The remaining eight OVX mice were sacrificed as above at 2
months following surgery (OVX 2). The left tibiae were obtained and
fixed in 4% paraformaldehyde for micro-computer tomography (CT)
analysis, while the right tibiae were immediately snap-frozen in
liquid nitrogen for RNA (miRNA and mRNA) isolation.
Micro-CT analysis
After being fixed with 4% paraformaldehyde for 48 h,
the left tibiae of each mouse were washed with phosphate-buffered
saline (PBS) three times and stored in 70% ethanol prior to
scanning. The fixed tibiae were then analyzed using a
high-resolution micro-CT (Skyscan 1072; Skyscan, Aartselaar,
Belgium). The scanning protocol was set at an isometric resolution
of 9 µm, and X-ray energy settings of 70 kV and 80
µA. After reconstruction, the region of interest of the
tibia was set at 0.5 mm from the proximal growth plate and selected
for further qualitative and quantitative analysis. Several
structural parameters, including trabecular bone volume per total
volume (BV/TV), mean trabecular thickness (Tb.Th), mean trabecular
number (Tb.N) and mean trabecular separation (Tb.Sp) were measured
by micro-CT as reported previously (16).
Microarray analysis
Total RNA was isolated from BMMs that had been
treated with RANKL for 0 or 5 days using the RNeasy®
Mini Kit (Invitrogen; Thermo Fisher Scientific, Inc.). The samples
were then submitted to LC Sciences (Houston, TX, USA) for
microarray analysis. The probe content was consistent with the
miRBase database version 14.0 (http://www.mirbase.org/). The fluorescence value was
determined by subtracting the background. The ratio of the two
subgroups (log2 transformed) and P-values were calculated using
Student's t-test. MiRNAs with P<0.01 and fluorescence values
>500 were selected.
Reverse-transcription quantitative
polymerase chain reaction (RT-qPCR) analysis
Homogenized tissue was obtained by grinding with
liquid nitrogen. Total RNA (including miRNA) was isolated from
cells and tissues by using the RNeasy® Mini kit
(Invitrogen; Thermo Fisher Scientific, Inc.) and reverse
transcription of 1.0 µg total RNA was performed using the
miRNA cDNA kit or the HiFiScript cDNA kit (CWBIO, Beijing, China),
which were used to analyze the expression levels of miRNA and
osteoclast-specific genes, respectively. Amplification reactions
were set up in 20-µl reaction volumes containing
amplification primers and UltraSYBR Mixture (with ROX) (CWBIO), and
products were detected using an ABI 7500 sequencing detection
system (Thermo Fisher Scientific, Inc.). A 1-µl volume of
cDNA and 1-µl of primers (Sangon Biotech Co., Ltd.,
Shanghai, China) were used in each amplification reaction. The
conditions of the reaction were as follows: Activation at 95°C for
10 min; 40 cycles of amplification, 95°C for 10 sec, 60°C for 24
sec and 72°C for 20 sec; and final extension at 72°C for 1 min. All
reactions were run in triplicate and were normalized to the miRNA
house-keeping gene U6, or mRNA house-keeping gene 18S and
glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Expression was
quantified using the ΔCq method, as described previously (17).
Primer sequences were as follows: 18S forward,
5′-CCTGCGGCTTAATTTGACTC-3′ and reverse, 5′-AACTAAGAACGGCCATGCAC-3′;
GAPDH forward, 5′-ACCCAGAAGACTGTGGATGG-3′ and reverse,
5′-CACATTGGGGGTAGGAACAC-3′; TRAP forward,
5′-CCATTGTTAGCCACATACGG-3′ and reverse, 5′-CACTCAGCACATAGCCCACA-3′;
Ctsk forward, 5′-TCCGCAATCCTTACCGAATA-3′ and reverse,
5′-AACTTGAACACCCACATCCTG-3′; NFATc1 forward,
5′-TCCACCCACTTCTGACTTCC-3′ and reverse, 5′-CTTCGCCCACTGATACGAG-3′;
U6 forward, 5′-CTCGCTTCGGCAGCACA-3′ and reverse,
5′-AACGCTTCACGAATTTGCGT-3′; mmu-miR-27 forward,
5′-GTGGTTCACAGTGGCTAAG-3′; mmu-miR-21a-5p forward,
5′-GTTTGGTAGCTTATCAGACTGA-3′; mmu-let-7i-5p forward,
5′-GTTTGGTGAGGTAGTAGTTTGT-3′; mmu-miR-22-3p forward,
5′-GTGAAGCTGCCAGTTGAAG-3′; mmu-miR-378-3p forward,
5′-GTTGACTGGACTTGGAGTC-3′; mmu-miR-340-5p forward,
5′-GTGGGGTTATAAAGCAATGAGA-3′; mmu-miR-23a-5p forward,
5′-TTGGGGGTTCCTGGGGAT-3′; common reverse, GTGCAGGGTCCGAGGT.
Statistical analysis
GraphPad Prism® software (GraphPad Inc.,
La Jolla, CA, USA) was used for all the statistical analyses.
Values are expressed as the mean ± standard error of the mean.
Analysis of variance followed by Dunnett's post hoc test was used
for comparisons between groups. All the experiments were performed
at least three times and P<0.05 was considered to indicate a
statistically significant difference.
Results
Validation of osteoclastogenesis in
vitro
In order to obtain miRNAs during various stages of
osteoclast differentiation, BMMs were treated with M-CSF and RANKL
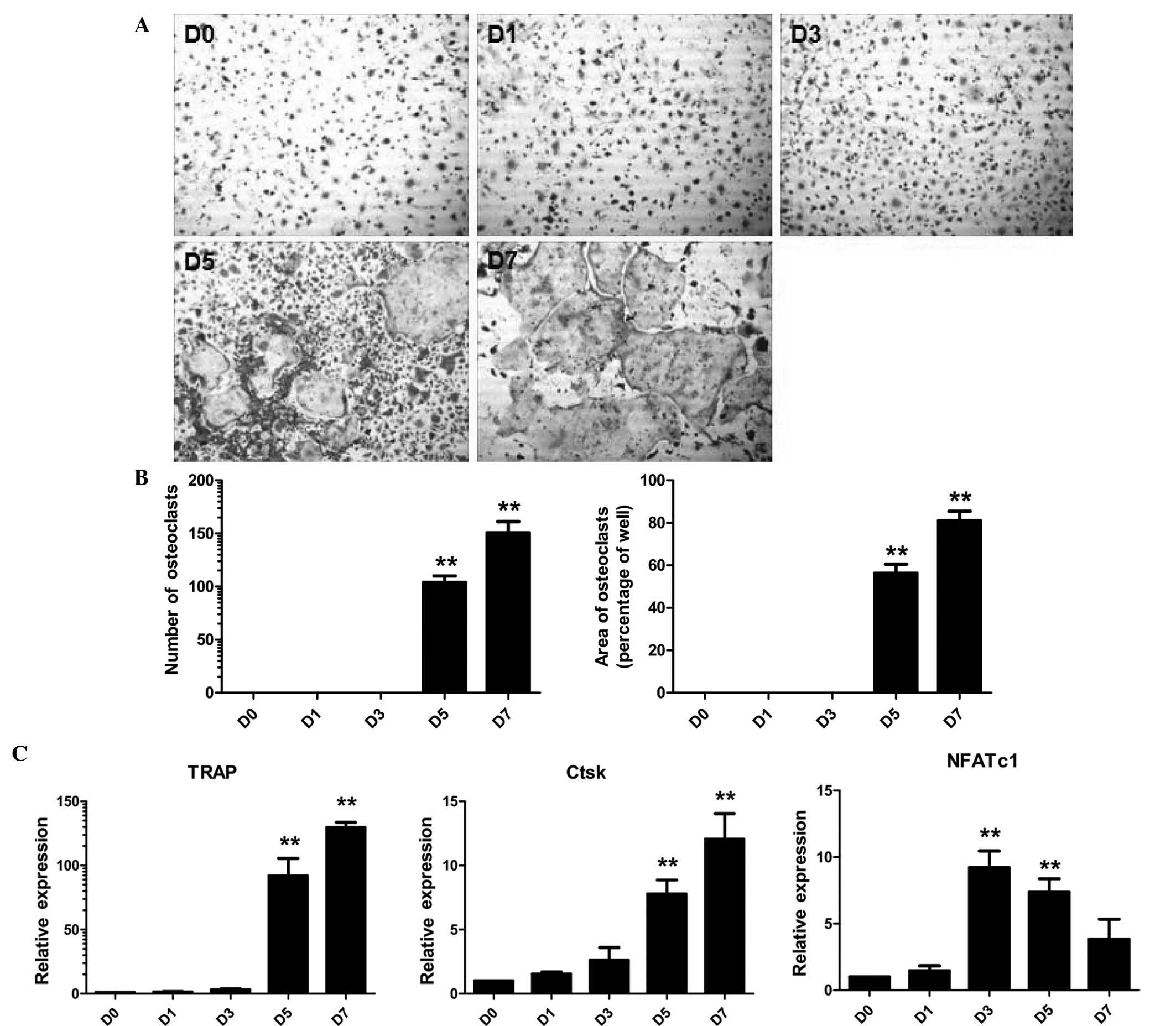
for 0, 1, 3, 5 or 7 days. As presented in Fig. 1A, BMMs were induced into mature
osteoclasts following treatment with RANKL for 5 or 7 days. While
no osteoclasts were present at days 0–3, the number of osteoclasts
(TRAP-positive cells with >3 nuclei) was 103.93±3.55 per well
and 150.67±6.13 per well following treatment of RANKL for 5 and 7
days, respectively (Fig. 1B).
Furthermore, the area of osteoclasts was 56.30±2.43
µm2 and 81.03±2.56 µm2,
respectively, following treatment with RANKL for 5 and 7 days
(Fig. 1B). In addition, the
expression levels of osteoclast-specific genes Ctsk, NFATc1 and
TRAP were also assessed. As expected, the osteoclast-associated
genes were upregulated in response to stimulation with RANKL.
NFATc1 levels peaked at day three, while the expression of TRAP and
Ctsk was low until day 3 and significantly increased on day 5 with
a further increase on day 7 (Fig.
1C). These results suggested that osteoclast maturation follows
stimulation with RANKL for 5–7 days.
Downregulated miRNAs during osteoclast
differentiation
Based on the successful induction of osteoclasts,
the expression of miRNAs during osteoclastogenesis was investigated
using total RNA extracted from BMMs (day 0) and mature osteoclasts
(day 5) using microarray analysis. As listed in Table I, six miRNAs that were
downregulated during osteoclastogenesis with a fold change of
>1.5 were identified, including miR-340-5p, miR-23a-5p,
miR-22-3p, miR-21-5p, miR-27a-3p and let-7i-5p. Largely consistent
with the results of previous studies, the downregulation of further
miRNAs, including miR-21, miR-155, miR-223 and miR-146a, was
identified by microarray analysis (presented in Table II) (11,12,18–27).
 | Table IMiRs with fold-changes of >1.5
during osteoclastogenesis between days 0 and 5 according to
microarray analysis. |
Table I
MiRs with fold-changes of >1.5
during osteoclastogenesis between days 0 and 5 according to
microarray analysis.
| Probe ID | Fold change (day 5
vs. 0) | Standard
deviation |
|---|
| mmu-miR-22-3p | 1.53 | 0.13 |
| mmu-miR-21-5p | 2.02 | 0.23 |
| mmu-miR-27a-3p | 1.57 | 0.35 |
| mmu-let-7i-5p | 1.64 | 0.33 |
| mmu-miR-340-5p | 1.70 | 0.18 |
| mmu-miR-23a-5p | 3.87 | 0.08 |
 | Table IImiRs with a known function in
osteoclasts and aberrant expression during ostoclastogenesis. |
Table II
miRs with a known function in
osteoclasts and aberrant expression during ostoclastogenesis.
| miR | Function | Expression
| Reference |
|---|
| Present study | Fold change (day 5
vs. 0) | Previous study |
|---|
| miR-223 | Suppression of
NFI-A | Downregulated | 0.191 | Downregulated | 18 |
| miR-155 | Suppression of MITF
and SOCS1 | Downregulated | 0.017 | Downregulated | 19 |
| miR-296 | Suppression of
c-Fos and MMP2 | Downregulated | 0.756 | Downregulated | 11 |
| miR-21 | Suppression of
PDCD4 | Downregulated | 0.494 | Upregulated | 12 |
| miR-146a | Suppression of
TRAF6 | Downregulated | 0.351 | Downregulated | 20 |
| miR-133a | Suppression of MITF
and MMP14 | No change | 1.000 | Downregulated | 21 |
| miR-219 | Suppression of MITF
and TRAF6 | No change | 1.000 | Downregulated | 21 |
| miR-148a | Suppression of
MAFB | Upregulated | 2.178 | Upregulated | 22 |
| miR-124 | Suppression of
NFATc1 | No change | 1.000 | Downregulated | 23 |
| miR-31 | Suppression of
RhoA | No change | 1.000 | Upregulated | 24 |
| miR-503 | Suppression of
RANK | Downregulated | 0.030 | Downregulated | 25 |
| miR-125a | Suppression of
TRAF6 | Downregulated | 0.772 | Downregulated | 26 |
| miR-141 | Suppression of MITF
and CALCR | No change | 1.000 | Downregulated | 21 |
| miR-190 | Suppression of
CALCR | No change | 1.000 | Downregulated | 21 |
| miR-34a | Suppression of
Tgif2 | Downregulated | 0.801 | Downregulated | 27 |
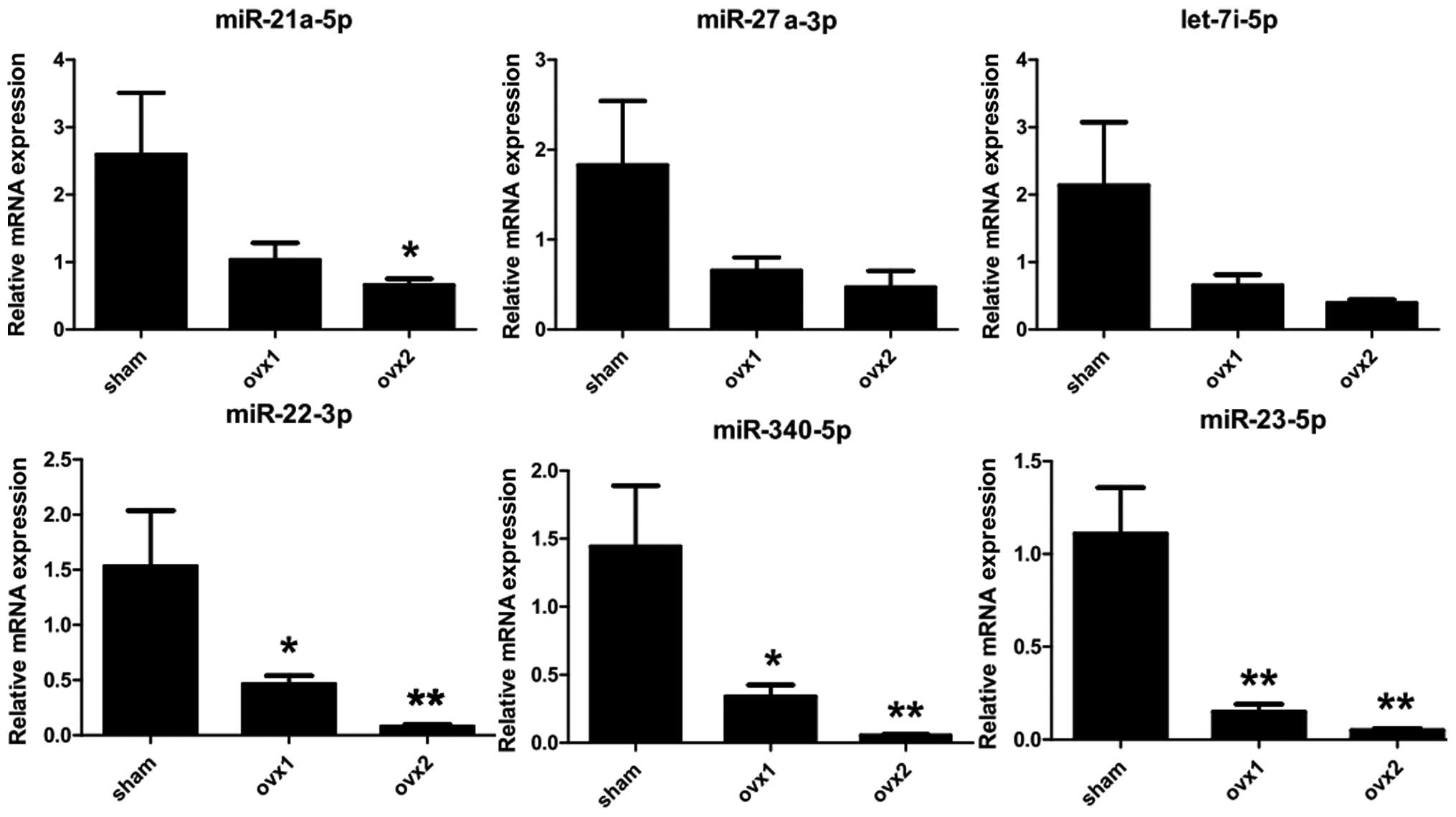
Furthermore, changes in the expression of the six
aberrantly expressed miRNAs during osteoclastogenesis was confirmed
using RT-qPCR. As presented in Fig.
2, miR-21a-5p, miR-27a-3p, let-7i-5p, miR-22-3p, miR-340-5p and
miR-23a-5p were observed to be downregulated during osteoclast
differentiation. The expression of miR-27a-3p, miR-21a-5p and
miR-23a-5p in matured osteoclasts was demonstrated to be ~70–80%
lower than that in BMMs without RANKL stimulation, while the
expression of let-7i-5p and miR-22-3p dropped to 50~70% in BMMs
following treatment with RANKL for 5 or 7 days. Notably, the
expression profiles of these miRNAs demonstrated a time-dependent
decrease during osteoclast formation. These results confirmed the
downregulation of a number of miRNAs during osteoclastogenesis.
Generation of an OVX-induced osteoporosis
model with various degree of bone loss
An OVX-induced murine osteoporosis model was used to
establish various degrees of osteoporosis in vivo.
Microarchitectural changes of the tibiae were then detected by
micro-CT (Fig. 3A). In accordance
with other studies (21), OVX mice
showed lower BV/TV and Tb.N, and higher Tb.Sp compared to
sham-operated mice (Fig. 3B). In
addition, the mice sacrificed two months after ovariectomy (OVX 2
group) displayed a reduced bone volume compared with that of the
mice in the OVX 1 group, with BV/TVs of 7.89±0.27 and 4.60±0.44%,
respectively (Fig. 3B). In
addition, OVX 2 mice exhibited a lower Tb.N and a higher Tb.Sp than
the OVX 1 mice (Fig. 3B). The
Tb.Th value remained constant indicating that OVX-induced
osteoporosis occurred predominantly by decreasing BV/TV and Tb.N.
The changes in the factors of bone intactness indicated that the
degree of osteoporosis in OVX 2 mice was higher than that in OVX 1
mice.
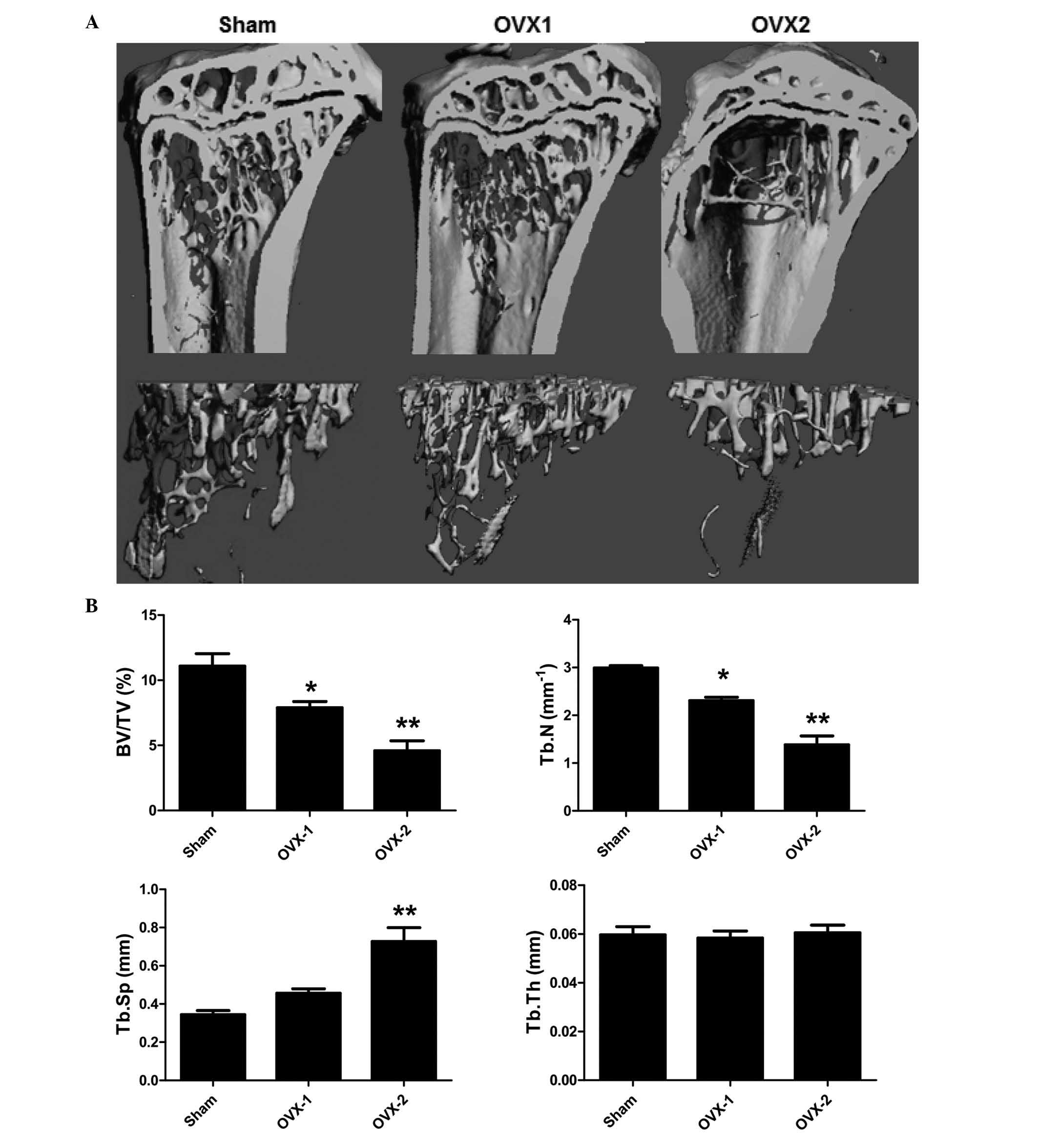 | Figure 3Generation of an OVX-induced
osteoporosis model with various degrees of bone loss. (A) The fixed
tibiae were analyzed by micro-computed tomography and
three-dimensionally reconstructed images from each group are shown
(magnification, ×40). (B) For each sample, BV/TV, Tb.Th, Tb.N and
Tb.Sp were measured. Values are expressed as mean ± standard error
of the mean. *P<0.05, **P<0.01 vs. the
sham group. BV, trabecular bone volume; TV, total volume; Tb.Th,
mean trabecular thickness; Tb.N, mean trabecular number; Tb.Sp,
mean trabecular separation; OVX1, mice at 1 month following OVX;
OVX2, mice at 2 months following OVX; OVX, ovariectomy. |
Osteoclastogenesis-associated miRNAs are
aberrantly expressed in OVX mice
Based on the expression of miRNAs during osteoclast
differentiation according to the microarray and RT-qPCR analyses of
the present study, the expression of these miRNAs was assessed in
the tibiae of a mouse model of osteoporosis. As shown in Fig. 4, miR-21a-5p, miR-27a-3p, let-7i-5p,
miR-22-3p, miR-340-5p and miR-23a-5p were found to be downregulated
in OVX 1 mice by at least 60% when compared to the sham-operated
mice. Of note, the expression of all of these miRNAs was further
declined in OVX 2 mice when compared to that in OVX 1 mice,
indicating a time-dependent decrease in the expression of these
miRNAs during the progression of osteoporosis.
Discussion
The present study used a microarray assay to
identify that miR-21a-5p, miR-27a-3p, let-7i-5p, miR-22-3p,
miR-340-5p and miR-23a-5p were downregulated during
osteoclastogenesis. This result was further supported by RT-qPCR
analysis of BMMs induced to undergo osteoclastogenesis by
stimulation with RANKL in vitro as well as in a murine model
of progressive osteoporosis in vivo. Of note, the expression
of these miRNAs was associated with the progression of
osteoporosis, with lower expression in more severe osteoporosis.
These miRNAs may represent potential biomarkers and targets for the
diagnosis and treatment of osteolytic diseases such as
osteoporosis.
However, the present study had several limitations.
First, osteoclasts are not the only type of cell in bone tissue.
The expression of miRNAs in the tibiae of the in vivo model
represents the expression levels in whole bone tissue, including
osteoclasts, osteoblasts and osteocytes. A more significant
decrease of these miRNAs was observed in vivo compared with
that obtained in vitro. Thus, it can be assumed that the
expression of these miRNAs may also have been altered in
osteoblasts and osteocytes during the development and progression
of osteoporosis.
The functional role of the miRNAs identified in the
present study in osteoclasts requires further elucidation. In
previous studies, the expression of miR-27a-3p was increased during
odontoblastic differentiation of hFOB1.19 as well as MDPC-23 cells
(28,29). By regulating the APC gene, miR-27
can affect β-catenin accumulation during osteoblast and odontoblast
differentiation (28). However,
whether the downregulation of miR-27 in osteoclasts is involved in
the wnt/β-catenin signaling pathway requires further research.
Similarly, miR-22 was associated with rheumatoid arthritis via a
novel p53/miR-22/cysteine-rich angiogenic inducer 61 axis in
synovial cells (30). In
hepatocytes, miR-22 significantly suppressed nuclear factor-κB
(NF-κB) activity by regulating the expression of nuclear receptor
co-activator 1 and nuclear receptor-interacting protein 1, two
NF-κB activators (31). However,
the regulation of NF-κB signaling by miR-22-3p during
osteoclastogenesis requires further investigation. In addition,
miR-340 was reported to regulate RAS/RAF/mitogen-activated protein
kinase (MAPK) signaling by modulating the expression of multiple
components of this pathway in melanoma (32). As the activation of the MAPK
pathway is crucial in RANKL-induced osteoclastogenesis, miR-340-5p
is likely to have a particular function in osteoclasts. In general,
further functional analysis of these miRNAs in osteoclasts is
required to reveal the underlying molecular mechanisms.
In conclusion, the present study identified six
aberrantly expressed miRNAs in BMMs undergoing osteoclastogenesis
and in mice with OVX-induced osteoporosis. The expression of these
miRNAs was shown to be associated with the degree of bone loss in
osteoporosis. The identified miRNAs miR-21a-5p, miR-27a-3p,
let-7i-5p, miR-22-3p, miR-340-5p and miR-23a-5p are therefore of
potential diagnostic and therapeutic value for osteolytic
diseases.
Acknowledgments
The present study was supported by the National
Nature Science Fund of China (nos. 81271971, 81271972 and
31270997).
References
|
1
|
Rachner TD, Khosla S and Hofbauer LC:
Osteoporosis: Now and the future. Lancet. 377:1276–1287. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kim MS, Magno CL, Day CJ and Morrison NA:
Induction of chemokines and chemokine receptors CCR2b and CCR4 in
authentic human osteoclasts differentiated with RANKL and
osteoclast like cells differentiated by MCP-1 and RANTES. J Cell
Biochem. 97:512–518. 2006. View Article : Google Scholar
|
|
3
|
Matsuo K and Irie N: Osteoclastosteoblast
communication. Arch Biochem Biophys. 473:201–209. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Dempster DW, Lambing CL, Kostenuik PJ and
Grauer A: Role of RANK ligand and denosumab, a targeted RANK ligand
inhibitor, in bone health and osteoporosis: A review of preclinical
and clinical data. Clin Ther. 34:521–536. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chong Y, Zhang J, Guo X, Li G, Zhang S, Li
C, Jiao Z and Shao M: MicroRNA-503 acts as a tumor suppressor in
osteosarcoma by targeting L1CAM. PLoS One. 9:e1145852014.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kloos W, Vogel B and Blessing E: MiRNAs in
peripheral artery disease-something gripping this way comes. Vasa.
43:163–170. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Schneider MR: MicroRNAs as novel players
in skin development, homeostasis and disease. Br J Dermatol.
166:22–28. 2012. View Article : Google Scholar
|
|
8
|
Jayaswal V, Lutherborrow M, Ma DD and Yang
YH: Identification of microRNA-mRNA modules using microarray data.
BMC Genomics. 12:1382011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sugatani T and Hruska KA: MicroRNA-223 is
a key factor in osteoclast differentiation. J Cell Biochem.
101:996–999. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bluml S, Bonelli M, Niederreiter B,
Puchner A, Mayr G, Hayer S, Koenders MI, van den Berg WB, Smolen J
and Redlich K: Essential role of microRNA-155 in the pathogenesis
of autoimmune arthritis in mice. Arthritis Rheum. 63:1281–1288.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Rossi M, Pitari MR, Amodio N, Di Martino
MT, Conforti F, Leone E, Botta C, Paolino FM, Del Giudice T,
Iuliano E, et al: miR-29b negatively regulates human osteoclastic
cell differentiation and function: Implications for the treatment
of multiple myeloma-related bone disease. J Cell Physiol.
228:1506–1515. 2013. View Article : Google Scholar
|
|
12
|
Sugatani T, Vacher J and Hruska KA: A
microRNA expression signature of osteoclastogenesis. Blood.
117:3648–3657. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Krzeszinski JY, Wei W, Huynh H, Jin Z,
Wang X, Chang TC, Xie XJ, He L, Mangala LS, Lopez-Berestein G, et
al: miR-34a blocks osteoporosis and bone metastasis by inhibiting
osteoclastogenesis and Tgif2. Nature. 512:431–435. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Seeliger C, Karpinski K, Haug AT, Vester
H, Schmitt A, Bauer JS and van Griensven M: Five freely circulating
miRNAs and bone tissue miRNAs are associated with osteoporotic
fractures. J Bone Miner Res. 29:1718–1728. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li H, Zhai Z, Liu G, Tang T, Lin Z, Zheng
M, Qin A and Dai K: Sanguinarine inhibits osteoclast formation and
bone resorption via suppressing RANKL-induced activation of NF-kB
and ERK signaling pathways. Biochem Biophys Res Commun.
430:951–956. 2013. View Article : Google Scholar
|
|
16
|
Bouxsein ML, Boyd SK, Christiansen BA,
Guldberg RE, Jepsen KJ and Müller R: Guidelines for assessment of
bone microstructure in rodents using micro-computed tomography. J
Bone Miner Res. 25:1468–1486. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Imai T, et al: Evaluation of reference
genes for accurate normalization of gene expression for real
time-quantitative PCR in Pyrus pyrifolia using different tissue
samples and seasonal conditions. PLoS One. 9:e864922014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Shibuya H, Nakasa T, Adachi N, Nagata Y,
Ishikawa M, Deie M, Suzuki O and Ochi M: Overexpression of
microRNA-223 in rheumatoid arthritis synovium controls osteoclast
differentiation. Mod Rheumatol. 23:674–685. 2013. View Article : Google Scholar
|
|
19
|
Zhang J, Zhao H, Chen J, Xia B, Jin Y, Wei
W, Shen J and Huang Y: Interferon-β-induced miR-155 inhibits
osteoclast differentiation by targeting SOCS1 and MITF. FEBS Lett.
586:3255–3262. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Nakasa T, Shibuya H, Nagata Y, Niimoto T
and Ochi M: The inhibitory effect of microRNA-146a expression on
bone destruction in collagen-induced arthritis. Arthritis Rheum.
63:1582–1590. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ell B, Mercatali L, Ibrahim T, Campbell N,
Schwarzenbach H, Pantel K, Amadori D and Kang Y: Tumor-induced
osteoclast miRNA changes as regulators and biomarkers of osteolytic
bone metastasis. Cancer Cell. 24:542–556. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cheng P, Chen C, He HB, Hu R, Zhou HD, Xie
H, Zhu W, Dai RC, Wu XP, Liao EY and Luo XH: miR-148a regulates
osteoclastogenesis by targeting V-maf musculoaponeurotic
fibrosarcoma oncogene homolog B. J Bone Miner Res. 28:1180–1190.
2013. View Article : Google Scholar
|
|
23
|
Lee Y, Kim HJ, Park CK, Kim YG, Lee HJ,
Kim JY and Kim HH: MicroRNA-124 regulates osteoclast
differentiation. Bone. 56:383–389. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Mizoguchi F, Murakami Y, Saito T, Miyasaka
N and Kohsaka H: miR-31 controls osteoclast formation and bone
resorption by targeting RhoA. Arthritis Res Ther. 15:R1022013.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chen C, Cheng P, Xie H, Zhou HD, Wu XP,
Liao EY and Luo XH: MiR-503 regulates osteoclastogenesis via
targeting RANK. J Bone Miner Res. 29:338–347. 2014. View Article : Google Scholar
|
|
26
|
Guo LJ, Liao L, Yang L, Li Y and Jiang TJ:
MiR-125a TNF receptor-associated factor 6 to inhibit
osteoclastogenesis. Exp Cell Res. 321:142–152. 2014. View Article : Google Scholar
|
|
27
|
Wu S, Huang S, Ding J, Zhao Y, Liang L,
Liu T, Zhan R and He X: Multiple microRNAs modulate p21Cip1/Waf1
expression by directly targeting its 3′untranslated region.
Oncogene. 29:2302–2308. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Park MG, Kim JS, Park SY, Lee SA, Kim HJ,
Kim CS, Kim JS, Chun HS, Park JC and Kim do K: MicroRNA-27 promotes
the differentiation of odontoblastic cell by targeting APC and
activating Wnt/β-catenin signaling. Gene. 538:266–272. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang T and Xu Z: miR-27 promotes
osteoblast differentiation by modulating Wnt signaling. Biochem
Biophys Res Commun. 402:186–189. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lin J, Huo R, Xiao L, Zhu X, Xie J, Sun S,
He Y, Zhang J, Sun Y, Zhou Z, et al: A novel p53/microRNA-22/Cyr61
axis in synovial cells regulates inflammation in rheumatoid
arthritis. Arthritis Rheumatol. 66:49–59. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Takata A, Otsuka M, Kojima K, Yoshikawa T,
Kishikawa T, Yoshida H and Koike K: MicroRNA-22 and microRNA-140
suppress NF-kB activity by regulating the expression of NF-kB
coactivators. Biochem Biophys Res Commun. 411:826–831. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Poenitzsch Strong AM, Setaluri V and
Spiegelman VS: microRNA-340 as a modulator of RAS-RAF-MAPK
signaling in melanoma. Arch Biochem Biophys. 563:118–124. 2014.
View Article : Google Scholar : PubMed/NCBI
|