Introduction
One of the most ubiquitous and important pathogens
worldwide is human cytomegalovirus (HCMV) (1–4).
HCMV can cause serious diseases in specific groups of individuals,
including those with compromised immunity and neonates. The viral
infection may lead to congenital malformations in newborns and
multi-organ involvement in patients with reduced immunity, with a
wide range of cell tropism in HCMV infection (5–7).
The HCMV genome is ~230 kb long and comprises 165
open reading frames in clinical strains (8). The HCMV RL13 open reading frame is
commonly 909 bp in length in Han clinical strains, one of the RL11
gene families reported in our previous study (9). HCMV RL13 encodes a virion envelope
glycoprotein, which locates spatially on the outer surface of the
viral envelope (10,11). The inhibitory effects of RL13 have
been shown with clinical isolates, as well as with the bacterial
artificial chromosome-cloned genome in fibroblasts and epithelial
cells (10,11). A previous study reported that
RL13-encoded protein can bind human IgG and may contribute to HCMV
immune evasion (12). Thus, more
detailed investigations are required to determine the mechanism by
which RL13 potentially targets unknown host proteins involved in
viral replication.
The present study aimed to characterize the
interaction and co-localization of RLI3 and nucleoside diphosphate
linked moiety X (nudix)-type motif 14 (NUDT14) and provide evidence
that this interaction may be involved in HCMV DNA replication. This
may elucidate the molecular mechanisms underlying HCMV diseases and
aid in indicating an appropriate target for effective therapeutic
agents.
Materials and methods
Cells and virus
Astrocytoma U373MG cells were provided by Professor
Songya Lv from State Key Laboratory of Virology, College of Life
Sciences, Wuhan University (Wuhan, China). Human embryonic lung
fibroblast MRC-5 cells (Shanghai Institute of Biochemistry and Cell
Biology, Shanghai, China), human embryonic kidney HEK293 cells
(Shanghai Institute of Cell Biology and Biochemistry) and the
astrocytoma U373MG cells were cultured in Dulbecco's modified
Eagle's medium (DMEM; GE Healthcare Life Sciences, Logan, UT, USA),
which was mixed with 10% fetal bovine serum (GE Healthcare Life
Sciences) at 37°C and 5% CO2. A low-passage clinical
isolate Han strain, which was isolated from a urine sample from a
congenitally HCMV-infected infant at the Department of Pediatrics
and maintained in the Virus Laboratory of The Affiliated Shengjing
Hospital, China Medical University (Shenyang, China) was propagated
in MRC-5 cells.
Yeast two-hybrid screening
The RL13 gene coding sequence was amplified by
polymerase chain reaction using HCMV Han strain DNA (Genbank no.
KJ426589.1; www.ncbi.nlm.nih.gov) as a template, and the following
primers, obtained from Invitrogen (Thermo Fisher Scientific, Inc.,
Waltham, MA, USA): Forward 5′-CCGGGCCATGGAGGCCAATAACACGTGCTCC-3′
and reverse 5′-CGCGGATCCGGTACCTTAGGTTTTAGTCCA-3′. The PCR
amplification was performed in a volume of 50 µl containing
3.5 µl DNA, 1 µl each primer, 5 µl 10X
TaqBuffer (with MgCl2), 2.5 µl dNTP mix (10 Mm),
1 unit Taq DNA polymerase (1 U/µl; Takara Biotechnology Co.,
Ltd., Dalian, China) and water up to 50 µl. The reaction was
performed in a Bio-Rad MyCycler Thermal Cycler (Bio-Rad
Laboratories, Inc., Hercules, CA, USA) and the reaction conditions
were as follows: 95°C for 5 min; 30 cycles of 95°C for 45 sec, 50°C
for 45 sec and 72°C for 1 min; followed by a final elongation at
72°C for 10 min. The PCR products were electrophoresed in 1.2%
agarose gel containing ethidium bromide (Sigma-Aldrich, St. Louis,
MO, USA) for visualization. The PCR products and the plasmids of
pGBKT7 vector (Clontech Laboratories, Inc., Mountain View, CA, USA)
were purified by wizard Genomic DNA Purification Kit (Promega
Corporation, Madison, WI, USA) in accordance with the
manufacturer's protocol. To construct the pGBKT7-RL13 plasmids,
enzyme digestion was performed by BamHI (Takara
Biotechnology Co., Ltd.) at 30°C for 2 h and SfiI (Takara
Biotechnology Co., Ltd.) at 50°C for 1 h. Following digestion, the
products were purified by wizard Genomic DNA Purification Kit
(Promega Corporation). Subsequently, the RL13 sequence was inserted
into the pGBKT7 vector (Clontech Laboratories, Inc) between the
SfiI and BamHI sites downstream of the c-Myc coding
sequence by T4 DNA ligase (Takara Biotechnology Co., Ltd.) and T4
DNA ligase buffer (Takara Biotechnology Co., Ltd.) at 16°C
overnight, resulting in pGBKT7-RL13 plasmids. The inserted sequence
of pGBKT7-RL13 was confirmed by sequencing (Invitrogen; Thermo
Fisher Scientific, Inc.). Yeast two-hybrid experiments were
performed in accordance with the manufacturer's protocol
(Matchmaker GAL4 Two-Hybrid System 3; Clontech Laboratories, Inc.).
The human fetus brain cDNA library was provided by Professor Gengfu
Xiao from the State Key Laboratory of Virology, College of Life
Sciences, Wuhan University, which was cloned into pACT2 (Clontech
Laboratories, Inc.), and the pGBKT7-RL13 were co-transformed into
the Saccharomyces cerevisiae strain, AH109 (Clontech
Laboratories, Inc.) by electroporation (Bio-Rad Laboratories, Inc.)
according to the protocol provided by the manufacturer. Positive
colonies were screened on synthetic dropout medium (Clontech
Laboratories, Inc.) in the absence of tryptophan, leucine, adenine
and histidine, and were detected using a chromogenic reaction of
x-α-Gal (Clontech Laboratories, Inc.) for α-galactosidase activity
(13). The plasmids containing
sequences encoding the interaction candidates of RL13, termed
pACT2-cDNA, were extracted and rescued via electrotransformation
into competent Escherichia coli DH5α (Clontech Laboratories,
Inc.). Each pACT2-cDNA plasmid was co-transformed into yeast with
either the pGBKT7-RL13 vector or the pGBKT7 empty vector. The
transformed cells were selected on QDO plates and underwent
assessment for β-galactosidase activity. Human gene sequences from
blue yeast colonies were sequenced (Invitrogen; Thermo Fisher
Scientific, Inc.), and analyzed using the BLAST database
(http://www.ncbi.nlm.gov/blast).
Glutathione S-transferase (GST) pull-down
experiments
The coding sequence of NUDT14, one of the candidate
proteins interacting with RL13 protein, was obtained from a
NUDT14-containing cDNA clone (pACT2-NUDT14), and was GST-tagged by
cloning into the pGEX-4T-2 vector (Clontech Laboratories, Inc.)
between the EcoRI and XhoI sites, yielding
pGEX-4T2-NUDT14.
A GST-pull-down experiment was performed, according
to the manufacturer's protocol (MagneGST™ Pull-Down System; Promega
Corporation). The c-Myc-labeled RL13 (c-Myc-RL13) was expressed
from pGBKT7-RL13 in a quick-coupled transcription/translation
reaction (TNT) T7 Quick Reaction (14). GST-labeled NUDT14 (GST-NUDT14) was
expressed in BL21 (DE3) cells (Tiangen Biotech Co., Ltd., Beijing,
China) transfected with pGEX-4T2-NUDT14 and induced with isopropyl
β-D-1-thiogalactopyranoside (IPTG; Clontech Laboratories, Inc.).
Subsequently, the MagneGST particles were incubated with the
reaction products for 30 min at room temperature. As a bait
protein, 20 µl of GST-NUDT14 was incubated with 80 µl
c-Myc-RL13 for 1.5 h on a rotating platform at room temperature.
Following washing with a GST binding/wash buffer (MagneGST™
Pull-Down System) three times, the bound proteins on the MagneGST
particles were eluted using elution buffer and solubilized in 2X
SDS sample buffer (Beyotime Institute of Biotechnology, Haimen,
China). The protein concentration was measured by ultraviolet
spectrophotometry as previously described (15). The binding proteins (20 mg) were
loaded in each lane of 12% polyacrylamide gels (SDS-PAGE;
Sigma-Aldrich) and the proteins were separated by gel
electrophoresis at a constant voltage of 100 V. The separated
proteins were transferred electrically onto nitrocellulose
membranes (Sigma-Aldrich), and then blocked with 5% bovine serum
albumin (BSA; Thermo Fisher Scientific, Inc.) for 2 h. Western blot
analyses were performed using monoclonal mouse anti-human
antibodies against c-Myc (1:1,000; Thermo Fisher Scientific, Inc.;
cat. no. AHO0052) or polyclonal goat anti-human antibodies against
GST monoclonal antibodies (1:1,000; Thermo Fisher Scientific, Inc.;
cat. no. PA5-18394) incubated overnight at 4°C and corresponding
goat anti-mouse peroxidase-conjugated secondary antibodies
(1:2,000; Beyotime Institute of Biotechnology; cat. no. A0216) and
rabbit anti-goat peroxidase-conjugated immunoglobulin G (IgG)
secondary antibodies (1:2,000; Absin Bioscience, Inc., Shanghai,
China; cat. no. abs20005) incubated for 2 h at room temperature.
The membranes were then reacted with the chemiluminescent
substrate, which was provided in the Western Chemiluminescent
Substrate kit (Pierce; Thermo Fisher Scientific, Inc.). Signals
were determined using a Molecular Imager ChemiDoc XRS System
(Bio-Rad Laboratories, Inc.). In the GST-pull-down assay, parallel
experiments were performed in controls containing GST- or
myc-tagged protein.
Co-immunoprecipitation analysis
The coding sequence of RL13 in pGBKT7-RL13 was
obtained and inserted into the pCMV-myc (Clontech Laboratories,
Inc.) between the SfiI and NotI sites, and designated
as pCMV-myc-RL13. The coding sequence of NUDT14 in pGEX-4T2-NUDT14
was obtained and cloned into the EcoRI and XhoI sites
of pCMV-HA (Clontech Laboratories, Inc.), and designated as
pCMV-HA-NUDT14. The constructs were confirmed by gene sequencing
(Invitrogen; Thermo Fisher Scientific, Inc.). The plasmids of
pCMV-myc-RL13 and pCMV-HA-NUDT14 were transiently co-transfected
into HEK293 cells using Lipofectamine 2000, according to the
manufacturer's protocol (Invitrogen; Thermo Fisher Scientific,
Inc.). The cell lysates were harvested with lysis buffer (M-PER™
Mammalian Protein Extraction Reagent; Thermo Fisher Scientific,
Inc.) supplemented with the protease inhibitors contained within
the Protein G Immunoprecipitation kit (Promega Corporation). The
protein concentration was determined using ultraviolet
spectrophotometry (15). The
co-immunoprecipitation analysis was performed, according to the
manufacturer's protocols of the ProFound Mammalian c-myc Tag
immunoprecipitation (IP)/Co-IP and hemagglutinin (HA) Tag IP/Co-IP
kits (Pierce; Thermo Fisher Scientific, Inc.). The binding proteins
(20 mg) were loaded in each lane of 12% polyacrylamide gels
(SDS-PAGE) and the proteins were separated by gel electrophoresis.
The separated proteins were electrotransferred onto nitrocellulose
membranes (Sigma-Aldrich, St. Louis, MO, USA), and then blocked
with 5% BSA (Thermo Fisher Scientific, Inc.) for 2 h. The
expression levels of myc-RL13 and HA-NUDT14 were detected using
Western blotting with mouse anti-human against myc or rabbit
anti-human polyclonal antibodies against HA (1:100; Thermo Fisher
Scientific, Inc., cat. no. 71-5500) incubated overnight at 4°C and
corresponding peroxidase-conjugated goat anti-mouse and goat
anti-rabbit (1:2,000; Beyotime Institute of Biotechnology; cat. no.
A0208) IgG secondary antibodies incubated at room temperature for 2
h. Imaging was performed using ChemiDoc™ XRS+ (Bio-Rad
Laboratories, Inc.) and density analysis was conducted with Image J
version 10.2 (National Institutes of Health, Bethesda, MD, USA).
Parallel co-immunoprecipitation experiments were performed with
specific tagged proteins containing myc or HA.
Cellular localization assay
To establish expression of the RL13 fusion protein
with a green fluorescent protein (EGFP) tag, the RL13 coding region
was amplified from the HCMV Han strain using the following primers:
Forward 5′-CGCTCGAGCCAATAACACGTGCTCC-3′ and reverse
5′-CGCGGATCCTTAGGTTTTAGTCCA-3′. Subsequently, the product was
inserted into the pEGFP-C1 (Clontech Laboratories, Inc.) via the
XhoI and BamHI sites, yielding pEGFP-C1-RL13.
Similarly, the coding sequence of NUDT14 in the NUDT14-containing
cDNA clone was fused to the Discosoma sp. red fluorescent
protein (DsRed) tag via amplification using PCR with the following
primers: Forward 5′-CGGAATTCCGTGTTGGTGAAGCAG-3′ and reverse
5′-CGCGGATCCGGAGCTATGCAAGCC-3′, and cloned into the pDsRed-C1
(Clontech) via the EcoRI and BamHI sites, yielding
pDsRed-C1-NUDT14. The constructs were confirmed by gene sequencing
(Invitrogen; Thermo Fisher Scientific, Inc.).
The HEK293 cells (5×106) were seeded into
a 35 mm confocal microscope dish (Nest Biotechnology Co., Ltd,
Jiangsu, China) 24 h prior to transfection. At 75% confluence, the
cells were transfected with either 4 µg pEGFP-C1-RL13, 4
µg pDsRed-C1-Nudt14, or a mixture of 2 µg
pEGFP-C1-RL13 and 2 µg pDsRed-C1-NUDT14 using Lipofectamine
2000 (Invitrogen; Thermo Fisher Scientific, Inc.), according to the
manufacturer's protocol. At 48 h post-transfection, the cells were
subjected to DAPI staining (Invitrogen; Thermo Fisher Scientific,
Inc.), and the expression levels of EGFP-RL13 and DsRed-NUDT14 were
detected using a laser scanning confocal microscope (Nikon Eclipse
C1 Plus; Nikon, Tokyo, Japan) with 488 and 543 nm excitation beams,
which corresponded to the EGFP-RL13 and DsRed-NUDT14 proteins,
respectively.
Stable expression of NUDT14 and
transfection of NUDT14-specific small interfering RNA (siRNA) into
cells
To generate a U373MG cell line, which stably
expressed NUDT14 (U373-S), the pDsRed-C1-NUDT14 vector and empty
vector, pDsRed-C1, were transfected into U373 cells using
Lipofectamine® LTX with Plus™ Reagent, according to the
manufacturer's protocol (Invitrogen; Thermo Fisher Scientific,
Inc.), yielding U373-S, and its control cell line, U373-C,
respectively. At 48 h post-transfection, neomycin (Thermo Fisher
Scientific, Inc.) was at a final concentration of 600 µg/ml
following mixing with culture medium. Neomycin-resistant cells were
screened with neomycin at ~2 weeks (16,17).
Protein extraction and quantification were performed as described
previously. Western blot analysis was used to analyze the
expression levels of NUDT14 in the selected cell clones, using
rabbit anti-human polyclonal antibodies against NUDT14 (1:1,000;
Abcam, Cambridge, MA, USA; cat. no. ab139656) and goat anti-rabbit
IgG antibody conjugated with horseradish peroxidase. Equal
quantities of sample were detected using western blotting, with
antibodies against NUDT14. The antibody was characterized and
specific bands for the NUDT14 protein were detected. As an internal
control, the expression level of cellular
glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was analyzed by
western blotting using mouse anti-human monoclonal antibody against
GAPDH (1:1,000; Abcam; cat. no. ab9482) and goat anti-mouse
antibody conjugated with horseradish peroxidase. The primary
antibodies were incubated overnight at 4°C and the secondary
antibodies were incubated for 2 h at room temperature.
The cells (n=1×105) were cultured and
transfected with Silencer® Select Pre-Designed NUDT14
siRNAs (Ambion; Thermo Fisher Scientific, Inc.) and control siRNA
(C-siRNA) (Ambion; Thermo Fisher Scientific, Inc.), respectively,
using Lipofectamine RNAiMAX Reagent, according to the
manufacturer's protocol (Invitrogen; Thermo Fisher Scientific,
Inc.). The antisense sequences of the NUDT14-specific siRNAs were:
5′-UUGAAUAAGAGAACGGUCAcg-3′ (S-siRNA-1) and
5′-AAAGAUGACGCCGAGGGUCtt-3′ (S-siRNA-2). In each well, 0.5
µl 10 nM siRNA and 3 µl Lipofectamine (Invitrogen,
Carlsbad, CA, USA) were diluted separately in 100 µl
Opti-MEM (Invitrogen; Thermo Fisher Scientific, Inc.). Following
incubation for 5 min at room temperature, the two solutions were
mixed. After 20 min, the mixture was added to the cells. At 10 h
post-transfection, the medium containing the siRNA was discarded,
and the transfection was repeated once to increase the transfection
efficiency and interference efficiency. At 24 h following the
secondary transfection, the cells were inoculated with the HCMV Han
strain at a multiplicity of infection of one. Total DNAs and
proteins were extracted from the infected cells 72 h following
infection, and the levels of NUDT14 in the infected cells were
determined using Western blot analysis.
Quantitative PCR (qPCR) analysis
In brief, DNA was extracted from the HCMV-infected
cells using a TIANamp Genomic DNA kit (Tiangen Biotech Co., Ltd.),
according to the manufacturer's protocol. The viral DNAs were
amplified and quantified using HCMV UL83-specific primers (forward
5′-GTCAGCGTTCGTGTTTCCCA-3′ and reverse 5′-GGGACACAACACCGTAAAGC-3′)
and a SYBR Green PCR Master Mix kit (Applied Biosystems; Thermo
Fisher Scientific, Inc.) on an ABI Prism 7300 Sequence Detection
System (Applied Biosystems; Thermo Fisher Scientific, Inc.). The
numbers of viral DNAs were normalized to the numbers of β-actin
detected in the same samples (using forward 5′-CGGAACCGCTCATTGCC-3′
and reverse 5′-ACCCACACTGTGCCCATCTA-3′ primers (18). The reaction mixture for qPCR
consisted of 4 µl DNA extract, 12.5 µl Power SYBR
Green PCR master mix (Applied Biosystems; Thermo Fisher Scientific,
Inc.), 0.5 µl of each primer at 10 µM, and an ABI
7300 device (Applied Biosystems; Thermo Fisher Scientific, Inc.)
was used. The amplification conditions were as follows: 95°C for 10
min, followed by 40 cycles of 95°C for 15 sec and 60°C for 1
min.
A modified comparative Cq method (2−ΔΔCq)
was used for relative quantification of viral DNAs (19–21).
The ΔCq values (Cqgene−Cqβ-Actin) were
calculated, following which the ΔΔCq (ΔCqtreated−ΔCqcontrol) were
calculated. The relative numbers of HCMV DNAs were described using
the equation 2−ΔΔCq (22). The experiments were repeated three
times.
Statistical analysis
Statistical significance was analyzed using analysis
of variance (ANOVA). P<0.05 was considered to indicate a
statistically significant difference. Data are presented as the
means ± standard deviation. All statistical analyses were computed
using SPSS (Version 13.0; SPSS, Inc., Chicago, IL, USA), and graphs
were produced using GraphPad Prism 5 (GraphPad Software, Inc., San
Diego, CA, USA).
Results
Identification of NUDT14 as a candidate
binding protein of HCMV RL13 using yeast two-hybrid screening
The candidate, pACT2-NUDT14, was screened to examine
its interaction with RL13. The transformation efficiency was almost
1.1×105 cfu/µg, and 14 yeast colonies produced
positive results. Sequencing and BLAST analysis indicated that 10
positive candidates interacted with the RL13 protein (Table I), one of which was 100% identical
to that of the human NUDT14 sequence in the National Center for
Biotechnology Information (Genbank no. NM177533.3).
 | Table IHomologous genes and homology were
analyzed by comparing the gene sequences of positive clones with
the human genome from Genebank. |
Table I
Homologous genes and homology were
analyzed by comparing the gene sequences of positive clones with
the human genome from Genebank.
| Sequence
number | Homologous
genes | Homology (%) |
|---|
| I | Homo sapiens
ankyrin repeat and GTPase domain Arf GTPase activating protein 11
(AGAP11) | 100 |
| II | Homo sapiens ligase
IV, DNA, ATP-dependent (LIG4) | 99 |
| III | Homo sapiens
tripartite motif containing 2 (TRIM2) | 99 |
| IV | Homo sapiens tweety
family member 3 (TTYH3) | 99 |
| V | Homo sapiens Sad1
and UNC84 domain containing 2 (SUN2) | 99 |
| VI | Homo sapiens
aspartate beta-hydroxylase domain containing 1(ASPHD1) | 100 |
| VII | Homo sapiens G
elongation factor, mitochondrial 2 (GFM2) | 98 |
| VIII | Homo sapiens ring
finger protein 19A, RBR E3 ubiquitin protein ligase (RNF19A) | 94 |
| IX | Homo sapiens leptin
receptor overlapping transcript-like 1(LEPROTL1) | 99 |
| X | Homo sapiens
nucleoside diphosphate linked moiety X (nudix)-type motif 14
(NUDT14) | 100 |
Identification of the interaction between
HCMV RL13 and host NUDT14 using a GST-pull-down assay
To further detect the binding between HCMV RL13 and
host NUDT14 in vitro, GST-tagged NUDT14 was used as a bait
protein and c-Myc-tagged RL13 was used as the prey protein in the
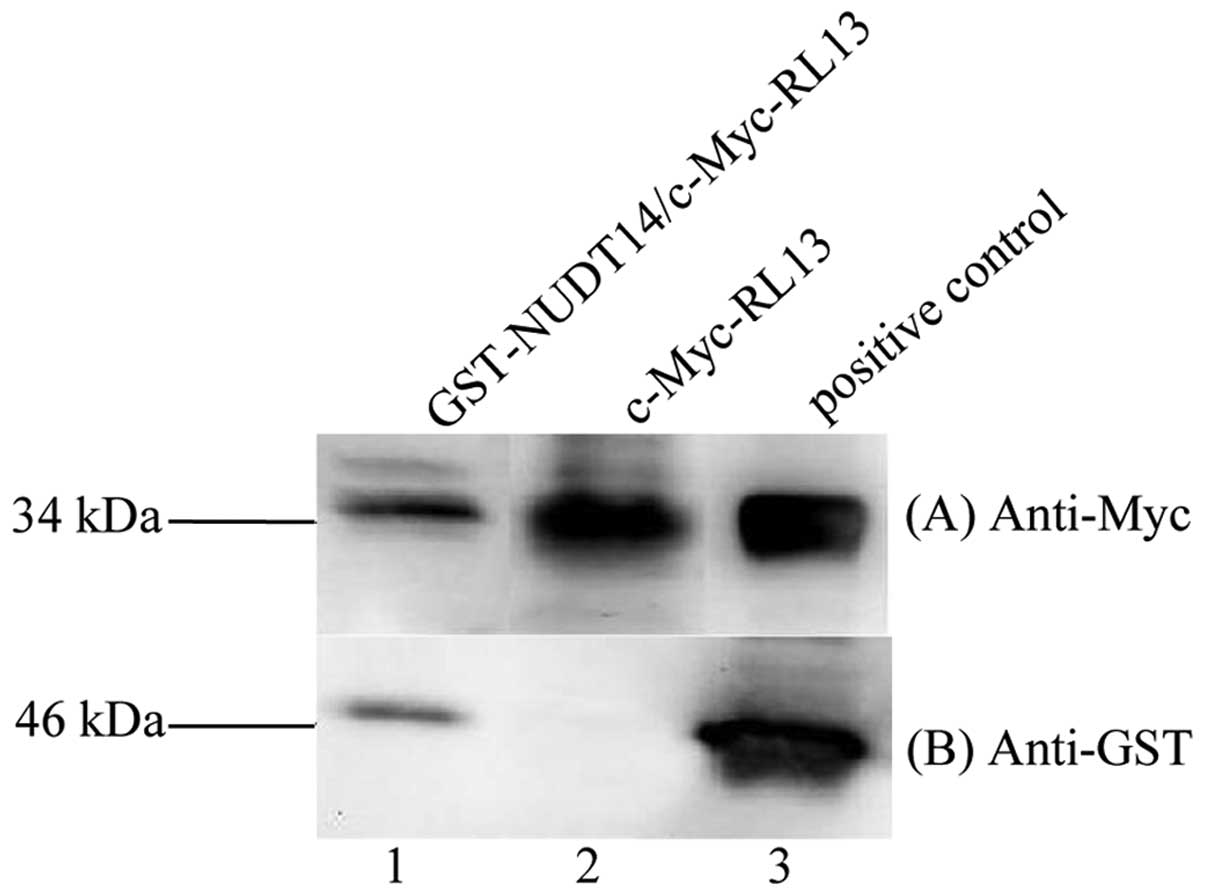
GST-pull down assay. As shown in Fig.
1, a c-Myc-labeled RL13 protein (~34 kDa) was captured, with a
GST-tagged NUDT14 protein of ~46 kDa using MagneGST particles, but
not with GST alone. These results showed that RL13 had the ability
to interact with NUDT14 in vitro.
Determination of the interaction between
HCMV RL13 and host NUDT14 by co-immunoprecipitation
To further confirm the interaction between RL13 and
NUDT14, a co-IP assay was performed. As shown in Fig. 2, the c-Myc-labeled RL13 and
HA-labeled NUDT14 proteins were assessed in the recovered products
following immunoprecipitation with either anti-c-Myc or anti-HA
antibodies (lanes 1 and 2, respectively). The input indicated that
the protein levels of c-Myc-labeled RL13 and HA-labeled NUDT14 in
the HEK293 cells were detected with anti-c-Myc and anti-HA
antibodies, respectively (Fig. 2;
lane 3). The positive control showed that the target proteins were
correct in position and size (Fig.
2; lane 4). These results confirmed the interaction between the
HCMV RL13 and host NUDT14 proteins in human cells.
RL13 and NUDT14 proteins are co-localized
in human cells by fluorescence confocal microscopy
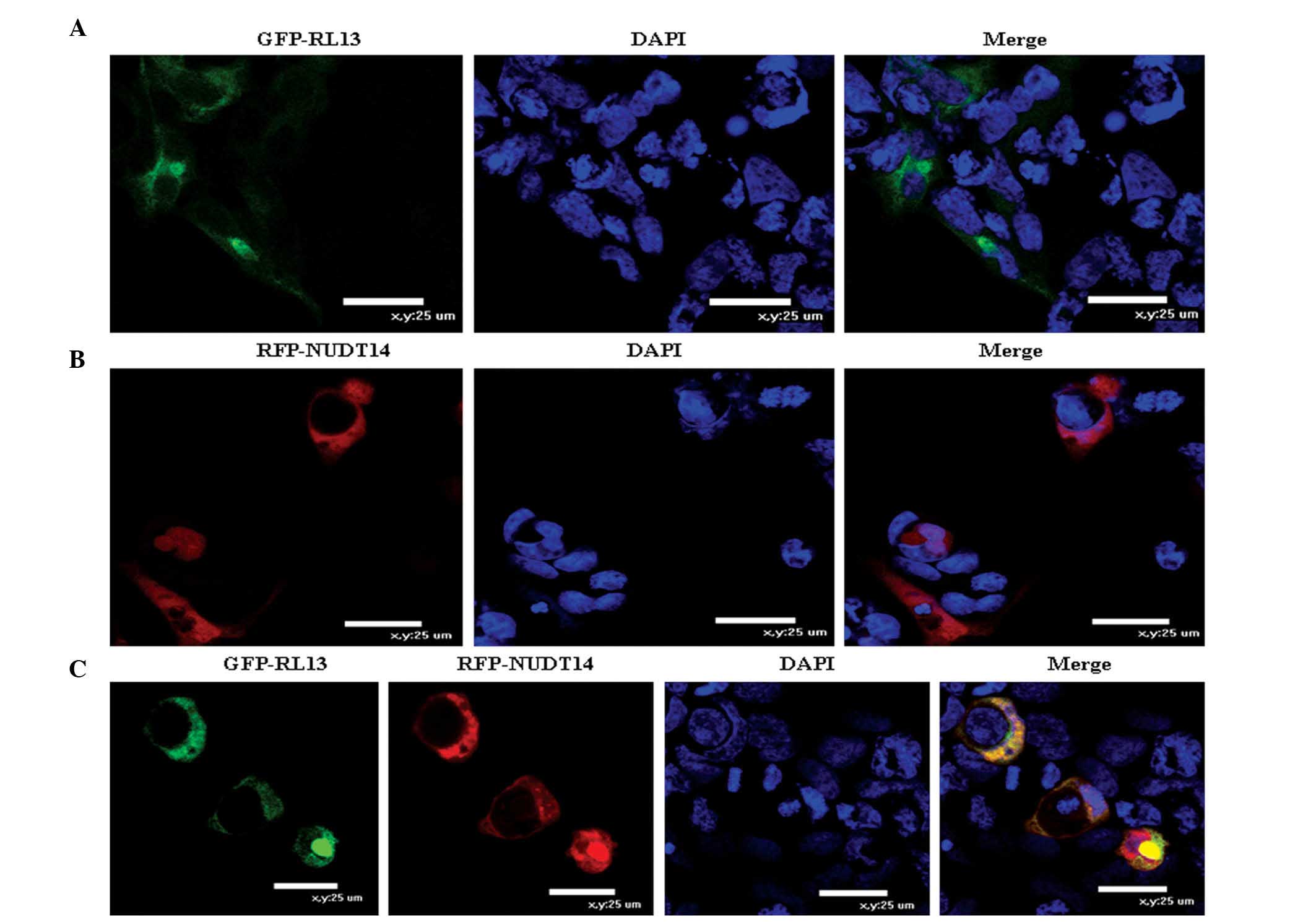
To determine whether the HCMV RL13 and NUDT14
proteins localized within the same cellular compartment, the HEK293
cells were transfected with pEGFP-RL13 (GFP-RL13), pDsRed-NUDT14
(RFP-NUDT14) or the two plasmids together, in the present study.
The resulting merged image represented regions of overlap among the
GFP-, RFP- and DAPI-stained images. The GFP-RL13 fusion proteins
were localized predominantly in the HEK293 cell membrane and
cytoplasm (Fig. 3A). Similarly,
The RFP-NUDT14 fusion proteins were predominantly localized in the
HEK293 cell membrane and cytoplasm (Fig. 3B). The fluorescent-labeled GPF-RL13
and RFP NUDT14 proteins were spatially co-localized and expressed
simultaneously in the HEK293 cell membrane and cytoplasm (Fig. 3C). Furthermore, the results showed
that the co-localization of the two proteins had no effects on the
distribution of either individually in the uninfected cells.
Effect of NUDT14 protein on HCMV DNA
replication
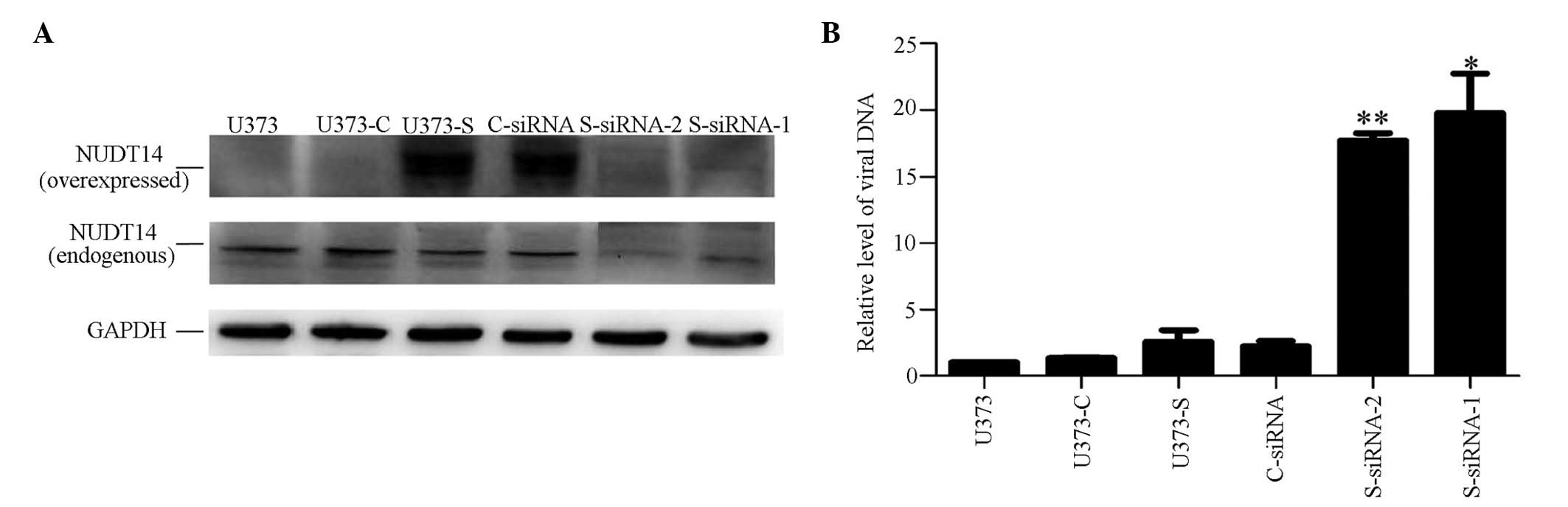
To investigate the effect of overexpressing NUDT14
on HCMV DNA replication, a stable NUDT14-expressing cell line,
U373-S, was constructed. No differences were observed between the
stable cell lines and the U373MG cells in growth characteristics or
survival. In the stable U373-S cell lines, the protein expression
of NUDT14 was five-fold higher than in the U373MG cells and the
empty vector-transfected cells (U373-C; Fig. 4A).
To investigate the effect of underexpressing NUDT14
on HCMV DNA replication, the U373-S cells were transfected with
NUDT14-specific siRNA molecules (S-siRNA). Compared to the U373-S
cells, the underexpression of NUDT14 mediated by specific siRNA
showed no effect on cell growth during the investigation. The
results of the Western blotting results showed that the protein
expression of NUDT14 in the S-siRNA-1- and S-siRNA-2-transfected
cells collected 72 h post-HCMV infection were significantly
reduced, by 84.14%, compared with the C-siRNA-transfected cells
(Fig. 4A).
To determine whether changes in the expression of
NUDT14 altered HCMV DNA replication, viral DNA copies were detected
using qPCR. At 72 h post-infection, no significant difference in
the relative copy number of HCMV, namely UL83/β-actin, in the
U373-S cells stably expressing NUDT14, relative to those in the
U373-C, U373MG and C-siRNA cells. However, following a decrease in
the expression of NUDT14, there was a 20-fold increase in the
number of HCMV copies in the infected cells treated with the two
NUDT14-specific S-siRNA-1 (ANOVA P=0.036) and S-siRNA-2 (ANOVA
P=0.002), compared with those in the cells transfected with C-siRNA
and the U373-S cells. The fact that the S-siRNA-1 and S-siRNA-2
molecules exhibited the same phenotype indicated that there was no
off-target of the siRNAs in the RNA interference experiments
(Fig. 4B). These results suggested
that inhibiting the expression of NUDT14 in the HCMV-infected cells
increased viral replication.
Discussion
Replication of herpes virus DNA is regarded as a
complex and controlled event during lytic infection (23,24).
Understanding the interaction between the virus and human host is
pivotal for clarifying the viral replication mechanism. Stanton
et al indicated that RL13 exerted independent, suppressive
effects on HCMV growth in fibroblasts, as well as epithelial cells
(10,11). However, the mechanism remains to be
elucidated.
In the present study, in order to investigate novel
human proteins, which potentially interact with the HCMV RL13
protein for elucidating the viral replication mechanism, a yeast
two-hybrid screen was performed. The results showed that RL13
interacted with 10 candidates, one of which was human NUDT14
(Table I). As a member of the
Nudix hydrolase family, NUDT14 encodes a UDP-glucose
pyrophosphatase (UGPPase), which reversibly catalyzes the formation
of UDP-glucose and pyrophosphate from UTP and glucose 1-phoshpate
in the presence of Mg2+ (25). To further identify the potential
interaction, a GST pull-down experiment was performed. As shown in
Fig. 1, c-Myc-RL13 was
specifically bound to GST-NUDT14 in vitro. To examine the
interaction between the RL13 protein and NUDT14 protein in more
detail, a co-immunoprecipitation experiment using HEK293 cells was
performed. As shown in Fig. 2,
c-Myc-RL13 was specifically immunoprecipitated with HA-NUDT14. To
further characterize the interaction, co-localization of these two
proteins was confirmed by confocal microscopy analysis. As shown in
Fig. 3, GFP-RL13 and RFP-NUDT14
were spatially co-localized in the HEK293 cells. These results
provided precise evidence that the RL13 protein interacted with the
NUDT14 protein in mammalian cells. The present study is the first,
to the best of our knowledge, to report on the interactions between
any HCMV proteins and NUDT14 proteins.
NUDT14 encodes a UGPPase, which belongs to the Nudix
hydrolases superfamily with versatile, Mg2+-requiring,
'housecleaning' characteristics, and may exert housecleaning
functions in eliminating toxic metabolites (26,27).
It has been reported that UGPPase may function in modulating
glycogen metabolism, and may be vital in connecting the
gluconeogenic process with other metabolic pathways to meet the
physiological and biochemical requirements of the mammalian cell
(28–31). The results of the present study
suggested s specific interaction between NUDT14 and RL13 in the
cell membrane and cytoplasm, and further investigations may
determine whether this interaction affects HCMV DNA replication,
for the development of antiviral strategies to prevent HCMV
infection and disease. In reference to the above results, the
present study investigated whether the RL13 and NUDT14 proteins are
involved in viral replication.
In the present study, the underexpression of NUDT14
by a specific siRNA in HCMV-infected cells resulted in an increase
in the level of viral DNA (Fig.
4). UGPPase may exert a housecleaning role in eliminating toxic
metabolites, as a member of the nudix hydrolases (26,27).
As a specific type of toxic metabolite, HCMV infection may be
eliminated by cellular UGPPase. The underexpression of NUDT14 by
specific siRNA in HCMV-infected cells may impair housecleaning by
NUDT14, which may benefit viral DNA replication.
The present study provided additional evidence that
the overexpression of NUDT14 had no significant effects on host
cell growth or viral DNA replication in the U373-S cell line. This
may be due to the level of endogenous NUDT14 being sufficient to
maintain the normal physiological function of cells. This
hypothesis requires further experimental verification, as only the
interaction between HCMV RL13 and host NUDT14, and the effects of
altering the expression of NUDT14 in U373MG cells on viral DNA
replication, were confirmed in the present study. Whether changes
in the expression of NUDT14 in other HCMV-permissive cells affects
viral DNA replication remains to be elucidated.
In conclusion, the present study identified RL13 as
a late gene in HCMV lytic infection, shown by its interaction with
the host protein, NUDT14, and that inhibition of the expression of
cellular NUDT14 increased HCMV DNA replication. These findings
assist in beginning to understand the mechanisms of the interaction
between the HCMV RL13 protein and the NUDT14 host factor during
infection. An improved understanding of the interaction between
HCMV RL13 and host proteins may provide insight into the
fundamental cellular pathways involved in viral binding and entry
in lytic infections.
Acknowledgments
This study was jointly sponsored by a grant from the
National Natural Science Foundation of China (grant nos. 81171580
and 81171581) and the Outstanding Scientific Fund of Shengjing
Hospital (grant no. 201105).
References
|
1
|
Bissinger AL, Sinzger C, Kaiserling E and
Jahn G: Human cytomegalovirus as a direct pathogen: Correlation of
multiorgan involvement and cell distribution with clinical and
pathological findings in a case of congenital inclusion disease. J
Med Virol. 67:200–206. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Plachter B, Sinzger C and Jahn G: Cell
types involved in replication and distribution of human
cytomegalovirus. Adv Virus Res. 46:195–261. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sinzger C, Grefte A, Plachter B, Gouw AS,
The TH and Jahn G: Fibroblasts, epithelial cells, endothelial cells
and smooth muscle cells are major targets of human cytomegalovirus
infection in lung and gastrointestinal tissues. J Gen Virol.
76:741–750. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Mocarski ES: Cytomegaloviruses. Fields
Virology. Fields BN, Knipe DM and Howley PM: Lippincott-Raven;
Philadelphia, Pennsylvania, USA: pp. 2447–2492. 2006
|
|
5
|
Sinzger C, Plachter B, Grefte A, The TH
and Jahn G: Tissue macrophages are infected by human
cytomegalovirus in vivo. J Infect Dis. 173:240–245. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kahl M, Siegel-Axel D, Stenglein S, Jahn G
and Sinzger C: Efficient lytic infection of human arterial
endothelial cells by human cytomegalovirus strains. J Virol.
74:7628–7635. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Riegler S, Hebart H, Einsele H, Brossart
P, Jahn G and Sinzger C: Monocyte-derived dendritic cells are
permissive to the complete replicative cycle of human
cytomegalovirus. J Gen Virol. 81:393–399. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Dolan A, Cunningham C, Hector RD,
Hassan-Walker AF, Lee L, Addison C, Dargan DJ, McGeoch DJ, Gatherer
D, Emery VC, et al: Genetic content of wild-type human
cytomegalovirus. J Gen Virol. 85:1301–1312. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Li M, Ma Y, Ji Y, He R, Qi Y, Sun Z, Wang
N, Gao S and Ruan Q: Human cytomegalovirus RL13 gene transcripts in
a clinical strain. Virus Genes. 43:327–334. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Stanton RJ, Baluchova K, Dargan DJ,
Cunningham C, Sheehy O, Seirafian S, McSharry BP, Neale ML, Davies
JA, Tomasec P, et al: Reconstruction of the complete human
cytomegalovirus genome in a BAC reveals RL13 to be a potent
inhibitor of replication. J Clin Invest. 120:3191–3208. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Dargan DJ, Douglas E, Cunningham C,
Jamieson F, Stanton RJ, Baluchova K, McSharry BP, Tomasec P, Emery
VC, Percivalle E, et al: Sequential mutations associated with
adaptation of human cytomegalovirus to growth in cell culture. J
Gen Virol. 91:1535–1546. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cortese M, Calò S, D'Aurizio R, Lilja A,
Pacchiani N and Merola M: Recombinant human cytomegalovirus (HCMV)
RL13 binds human immunoglobulin G Fc. PLoS One. 7:e501662012.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
To A, Bai Y, Shen A, Gong H, Umamoto S, Lu
S and Liu F: Yeast two hybrid analyses reveal novel binary
interactions between human cytomegalovirus-encoded virion proteins.
PLoS One. 6:e177962011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
McMahon TP and Anders DG: Interactions
between human cytomegalovirus helicase-primase proteins. Virus Res.
86:39–52. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Stepanchenko NS, Novikova GV and Moshkov
IE: Protein quantification. Russ J Plant Physiol. 58:727–742. 2011.
View Article : Google Scholar
|
|
16
|
Liu F and Altman S: Inhibition of viral
gene expression by the catalytic RNA subunit of RNase P from
Escherichia coli. Genes Dev. 9:471–480. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Miller AD and Rosman GJ: Improved
retroviral vectors for gene transfer and expression. Biotechniques.
7:980–982. 984–986. 989–990. 1989.PubMed/NCBI
|
|
18
|
Hänfler J, Kreuzer Ka, Laurisch K, Rayes
N, Neuhaus P, Schmidt CA and Oettle H: Quantitation of
cytomegalovirus (hCMV) DNA and beta-actin DNA by duplex real-time
fluorescence PCR in solid organ (liver) transplant recipients. Med
Microbiol Immunol. 192:197–204. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Su J, Zhu Z, Xiong F and Wang Y: Hybrid
cytomegalovirus-U6 promoter-based plasmid vectors improve
efficiency of RNA interference in zebrafish. Mar Biotechnol (NY).
10:511–517. 2008. View Article : Google Scholar
|
|
20
|
Cahill AL, Moore JM, Sabar FI and Harkins
AB: Variability in RNA interference in neuroendocrine PC12 cell
lines stably transfected with an shRNA plasmid. J Neurosci Methods.
166:236–240. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
22
|
Duan QJ, Tao R, Hu MF and Shang SQ:
Efficient inhibition of human cytomegalovirus UL122 gene expression
in cell by small interfering RNAs. J Basic Microbiol. 49:531–537.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Mocarski ES, Shenk T and Pass RF:
Cytomegalovirus. Fields virology. Knipe DM, Howley PM, Griffin DE,
et al: Lippincott Williams & Wilkins; Philadelphia, PA: pp.
2701–2772. 2007
|
|
24
|
Roizman B, Knipe D and Whitley R: Herpes
simplex viruses. Fields virology. Knipe DM and Howley PM:
Lippincott Williams & Wilkins; Philadelphia, Pennsylvania, USA:
pp. 2503–2601. 2007
|
|
25
|
Aksamit RR and Ebner KE: Purification,
properties and kinetic analysis of UDP-glucose pyrophosphorylase
from bovine mammary tissue. Biochim Biophys Acta. 268:102–112.
1972. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Bessman MJ, Frick DN and O'Handley SF: The
MutT proteins or 'Nudix' hydrolases, a family of versatile, widely
distributed, 'housecleaning' enzymes. J Biol Chem. 271:25059–25062.
1996. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
McLennan AG: The nudix hydrolase
superfamily. Cell Mol Life Sci. 63:123–143. 2006. View Article : Google Scholar
|
|
28
|
Heyen CA, Tagliabracci VS, Zhai L and
Roach PJ: Characterization of mouse UDP-glucose pyrophosphatase, a
Nudix hydrolase encoded by the Nudt14 gene. Biochem Biophys Res
Commun. 390:1414–1418. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yagi T, Baroja-Fernández E, Yamamoto R,
Muñoz FJ, Akazawa T, Hong KS and Pozueta-Romero J: Cloning,
expression and characterization of a mammalian Nudix hydrolase-like
enzyme that cleaves the pyrophosphate bond of UDP-glucose. Biochem
J. 370:409–415. 2003. View Article : Google Scholar
|
|
30
|
Rodriguez-López M, Baroja-Fernández E,
Zandueta-Criado A and Pozueta-Romero J: Adenosine diphosphate
glucose pyrophosphatase: A plastidial phosphodiesterase that
prevents starch biosynthesis. Proc Natl Acad Sci USA. 97:8705–8710.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Moreno-Bruna B, Baroja-Fernández E, Muñoz
FJ, Bastarrica-Berasategui A, Zandueta-Criado A, Rodriguez-López M,
Lasa I, Akazawa T and Pozueta-Romero J: Adenosine diphosphate sugar
pyrophosphatase prevents glycogen biosyn-thesis in Escherichia
coli. Proc Natl Acad Sci USA. 98:8128–8132. 2001. View Article : Google Scholar
|