Introduction
With the rising incidence of type 2 diabetes
mellitus worldwide, particularly in developing countries (1), type 2 diabetes mellitus has become
one of the biggest mortality-causing diseases in humans. In
addition to its increasing incidence rate and the trend of
decreasing age at the onset, type 2 diabetes mellitus is a major
health problem, drawing attention worldwide. Type 2 diabetes
mellitus has high familial aggregation, and genetic and
environmental factors contribute to its incidence synergistically
(2). The genetic factors are
important in the pathogenesis process of type 2 diabetes (3). Numerous previous studies have
demonstrated that the type, number and nature of the mutations in
the alleles involved in the pathogenesis of type 2 diabetes may
exhibit significant differences among the different ethnic groups
or individuals. It was shown that even the different areas in the
same ethnic populations were differently correlation between gene
polymorphisms and type 2 diabetes. The present study was performed
to explore the pathological mechanisms of type 2 diabetes from the
genetic factors in order to prevent and treat type 2 diabetes more
effectively.
Hepatocyte nuclear factor (HNF)-4α is a highly
conservative member in the nuclear receptor superfamily, with the
characteristics of liposoluble hormone receptors. It is normally
expressed in the liver, kidney, intestine and pancreas (4,5), and
can directly enter into the nucleus to regulate the transcription
of genes involved in the regulation of the development,
differentiation and function of the islet β and liver cells, and
the maintenance of the glucose homeostasis (6). In 1996, HNF-4α was first identified
harboring the non-sense mutations of CAG (glutamine) to TAG (amber
mutation), termed the Q268X mutant, at +268 by a family linkage
analysis on a multigeneration of diabetes family. HNF-4α was
believed to be a disease-causing gene, termed Maturity-Onset
Diabetes of the Young 1 (7). In
subsequent studies, HNF-4α was also revealed to be associated with
type 2 diabetes (8,9). Previous data revealed that certain
genes causing single gene diabetes may also be associated with the
susceptibility of type 2 diabetes. The rare mutations of these
genes cause single gene diabetes mellitus, while the common
polymorphisms in these genes are associated with type 2 diabetes
(10). Previous studies also
identified that the location of HNF-4α genes in the chromosome area
20 q13.12 to 20 q13.13 is also a susceptible locus for type 2
diabetes (11) and has a greater
frequency of single nucleotide polymorphisms (SNPs). Therefore,
HNF-4α is a candidate gene for type 2 diabetes. However, whether
the incidence of type 2 diabetes is correlated with HNF-4α gene
polymorphisms remains to be elucidated.
The correlation between the gene polymorphisms in
HNF-4α and type 2 diabetes has been reported in the populations in
North America, Europe and East Asia. A total of four single
nucleotide polymorphisms, rs4810424, rs1884613, rs1884614 and
rs2144908, near the promoter of the HNF-4α gene in those
populations exhibited different correlations with type 2 diabetes
(12–16). However, little research on the
minorities in China exists. Yunnan province possesses >1 million
Bai individuals, who predominantly live in the Dali Bai autonomous
prefecture. The Bai nationality is the third biggest minority in
Yunnan province, harboring a number of patients with type 2
diabetes. Due to its geographical environment, social mores and the
characteristics of population migration, less intermarriage occurs
between different nationalities, leading to a relatively isolated
genetic population. Therefore, the present study was performed to
assess the correlation of the four SNPs, rs4810424, rs1884613,
rs1884614 and rs2144904, near the promoter of HNF-4α gene with the
patients type 2 diabetes in the Bai nationality. Additionally,
haplotype analysis was also performed to investigate if this can
increase the risk of type 2 diabetes in Dali Bai individuals.
Materials and methods
Patients and individuals
All individuals were of Dali Bai nationality.
Patients with type 2 diabetes were diagnosed strictly according to
the 1999 diagnostic criteria of diabetes from the World Health
Organization (WHO) (17). A total
of 44 hospitalized patients with type 2 diabetes in The People's
Hospital of Jianchuan County in Dali prefecture from 2010 to 2012,
including 20 males and 24 females, were enrolled in this study. The
patients were aged between 39 and 77 years. A total of 87 healthy
Bai individuals, aged between 35 and 70 years, were recruited as
the normal control group. The individuals in the normal control
group included 40 males and 47 females, without diabetes,
hypertension, coronary heart disease, high cholesterol or other
associated diseases. Written informed consent was obtained from
each individual and this study was performed in accordance with the
declaration of Helsinki. The present study approved by the Ethics
Committee of the Second Affiliated Hospital of Kunming Medical
University (Kunming, China).
Selection of SNPs
A total of four SNPs (rs4810424, rs1884613,
rs1884614 and rs2144904) near the promoter of the HNF-4α gene were
previously reported to be highly correlated with type 2 diabetes in
North America, Europe and East Asia. In the present study, these
SNPs were selected to assess their association with type 2 diabetes
in the Dali Bai individuals.
Detection of gene polymorphisms
DNA samples were extracted from the
EDTA-anticoagulant peripheral venous blood collected from each
individual. The DNA samples were used for polymerase chain reaction
(PCR) amplification, according to the manufacturer's instructions
in the DNA extraction kit [Tiangen Biotech (Beijing) Co., Ltd.,
Beijing, China] and PCR amplification kit (Sangon Biotech Co.,
Ltd., Shanghai, China). Specific primers for the HNF-4α gene, with
an enzyme digestion site for the four SNPs, were designed using
Premier 5.0 software (Premier Biosoft, Palo Alto, CA, USA)
(Table I) and were synthesized by
Sangon Biotech Co., Ltd. PCR-restriction fragment length
polymorphism (RFLP) was used to analyze the gene polymorphisms. The
PCR conditions were as follows: 3 min at 95°C, 35 cycles of 30 sec
at 94°C and 40 sec at 72°C, followed by 5 min at 72°C. Enzyme
digestion reactions were performed on the four PCR products at 37°C
for 10–12 h using the corresponding restriction enzymes
(SatI, Sau3AI, Alw441 and HaeIII; Bio
Basic, Inc., Markham, ON, Canada). The enzyme digestion products
were separated using 2% agarose gel electrophoresis and analyzed
using the Tanon 3500 Gel Imaging system (Tanon Science &
Technology Co., Ltd., Shanghai, China). The enzyme digestion
products were sequenced using the Sanger method (CapitalBio
Corporation, Beijing, China) to confirm the enzyme digestion
results.
 | Table ISpecific primers for the four SNP
sequences and the size of the products. |
Table I
Specific primers for the four SNP
sequences and the size of the products.
| SNP | Primer | Size of product
(bp) |
Tm(°C) | Enzyme recognition
site | Enzyme digestion
products (bp) |
|---|
| rs4810424 | F:
5′-GGGACTACAGGTGAATGCCA-3′ | 212 | 58 | SatI
(GC^NGC) | 212/127/85 |
| R:
5′-TCCTGTCTATTTCCCAGTGATTC-3′ |
| rs1884613 | F:
5′-TTGAGGACTCGTGATTCGCT-3′ | 113 | 56 | Sau3AI
(^GATC) | 153/114/39 |
| R:
5′-CAGTACCATGCAGTGGGGAT-3′ |
| rs1884614 | F:
5′-GTGTAACTTACCCAGAGGTGC-3′ | 117 | 56 | Alw44I
(G^TGCAC) | 157/117/40 |
| R:
5′-CAACTTTGGTTTTCACTTCTCA-3′ |
| rs2144908 | F:
5′-GTGGGGAACAAGGATGTAAAG-3′ | 111 | 56 | HaeIII
(GG^CC) | 151/109/42 |
| R:
5′-CCCTGATTCTGTCATTCCCT-3′ |
Statistical analysis
Hardy-Weinberg genetic balance of all subjects in
the type 2 diabetes group and normal control group were analyzed
using SHEsis software (http://analysis.bio-x.cn/SHEsisMain.htm). The
differences of the alleles and genotype frequencies between the two
groups were compared using Chi-square test, and subsequently the
Odds ratio and 95% Confidence interval were calculated. Haplotype
analysis was performed using SHEsis software. P<0.05 was
considered to indicate a statistically significant difference.
Results
Genotypes of rs4810424, rs1884613,
rs1884614 and rs2144904 detected by PCR-RFLP
The genotypes of the four SNPs (rs4810424,
rs1884613, rs1884614 and rs2144904) were analyzed using PCR-RFLP.
In rs4810424, the GC genotype possessed three bands (212/127/85
bp), the GG genotype only had one band (212 bp) and the CC genotype
had two bands (127 and 85 bp; Fig.
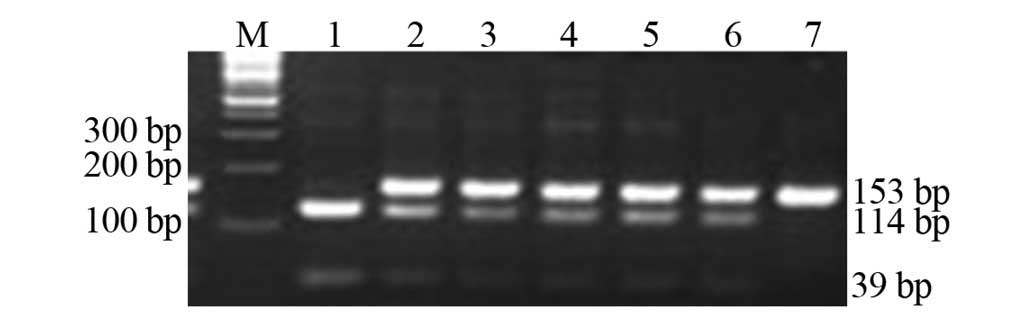
1). In rs1884613, three bands in the genotype GC (153/114/39
bp), two bands in the genotype GG (114 and 39 bp) and one band in
genotype CC (153 bp; Fig. 2) were
observed. As shown in Fig. 3, in
rs1884614, three bands represented the CT genotype (40/117/157 bp),
two bands represented the CC genotype (40/117 bp), and one band
represented the TT genotype (157 bp). In rs2144904, three
genotypes, AA (one band; 151 bp), GG (two bands; 42/109 bp) and AG
(three bands; 42/109/151 bp), were observed (Fig. 4). The sequencing results also
confirmed the genotyping results of enzyme digestion (Fig. 5).
Hardy-Weinberg equilibrium (HWE) balance
of the four SNPs in Dali Bai individuals
The four SNPs, rs4810424, rs1884613, rs1884614 and
rs2144904, near the promoter of the HNF-4α gene in the diabetes
group and the control group met the HWE law (P>0.05), meaning
that all of the four SNPs were in genetic equilibrium with a
population representative (Table
II).
 | Table IIHardy-Weinberg equilibrium results of
the four single nucleotide polymorphisms near the promoter of the
hepatic nuclear factor-4α gene in the diabetes group and the
control group. |
Table II
Hardy-Weinberg equilibrium results of
the four single nucleotide polymorphisms near the promoter of the
hepatic nuclear factor-4α gene in the diabetes group and the
control group.
| Group | rs4810424
| rs1884613
| rs1884614
| rs2144908
|
|---|
| χ2 | P | χ2 | P | χ2 | P | χ2 | P |
|---|
| T2DM | 0.44 | 0.51 | 0.051 | 0.82 | 1.01 | 0.32 | 1.01 | 0.32 |
| NC | 1.97 | 0.16 | 1.97 | 0.16 | 1.97 | 0.16 | 1.97 | 0.16 |
Correlation of the HNF-4α gene with type
2 diabetes mellitus
The frequencies of the alleles and genotypes of the
four SNPs near the promoter of the HNF-4α gene exhibited no
statistically significant differences between the diabetes group
and the control group in Dali Bai individuals. This suggested that
no association existed between rs4810424, rs1884613, rs1884614 and
rs2144908, and type 2 diabetes mellitus in Dali Bai individuals
(P>0.05; Table III).
 | Table IIIDistribution of the alleles and
genotypes of the four single nucleotide polymorphisms near the
promoter of hepatic nuclear factor-4α gene in Dali Bai
individuals. |
Table III
Distribution of the alleles and
genotypes of the four single nucleotide polymorphisms near the
promoter of hepatic nuclear factor-4α gene in Dali Bai
individuals.
| SNP | Group | Alleles [n (%)] | OR | P | Genotype [n (%)] | χ2 | P |
|---|
| | C | G | | | CC | CG | GG | | |
| rs4810424 | T2DM | 40 (45.5) | 48 (54.5) | 1.33 (0.79–2.24) | 0.28 | 8 (18.2) | 24 (54.5) | 12 (27.3) | 2.92 | 0.23 |
| NC | 67 (38.5) | 107 (61.5) | | | 16 (18.4) | 35 (40.2) | 36 (41.4) | | |
| | C | G | | | CC | CG | GG | | |
| rs1884613 | T2DM | 52 (59.1) | 36 (40.9) | 0.90
(0.54–1.53) | 0.71 | 15 (34.1) | 22 (50.0) | 7 (15.9) | 1.14 | 0.56 |
| NC | 107 (61.5) | 67 (38.5) | | | 36 (41.4) | 35 (40.2) | 16 (18.4) | | |
| | C | T | | | CC | CT | TT | | |
| rs1884614 | T2DM | 49 (55.7) | 39 (44.3) | 0.79
(0.47–1.32) | 0.37 | 12 (27.3) | 25 (56.8) | 7 (15.9) | 3.45 | 0.18 |
| NC | 107 (61.5) | 67 (38.5) | | | 36 (41.4) | 35 (40.2) | 16 (18.4) | | |
| | A | G | | | AA | AG | GG | | |
| rs2144908 | T2DM | 39 (44.3) | 49 (55.7) | 1.27
(0.76–2.14) | 0.37 | 7 (15.9) | 25 (56.8) | 12 (27.3) | 3.45 | 0.18 |
| NC | 67 (38.5) | 107 (61.5) | | | 16 (18.4) | 35 (40.2) | 36 (41.4) | | |
Haplotype analysis of the HNF-4α
gene
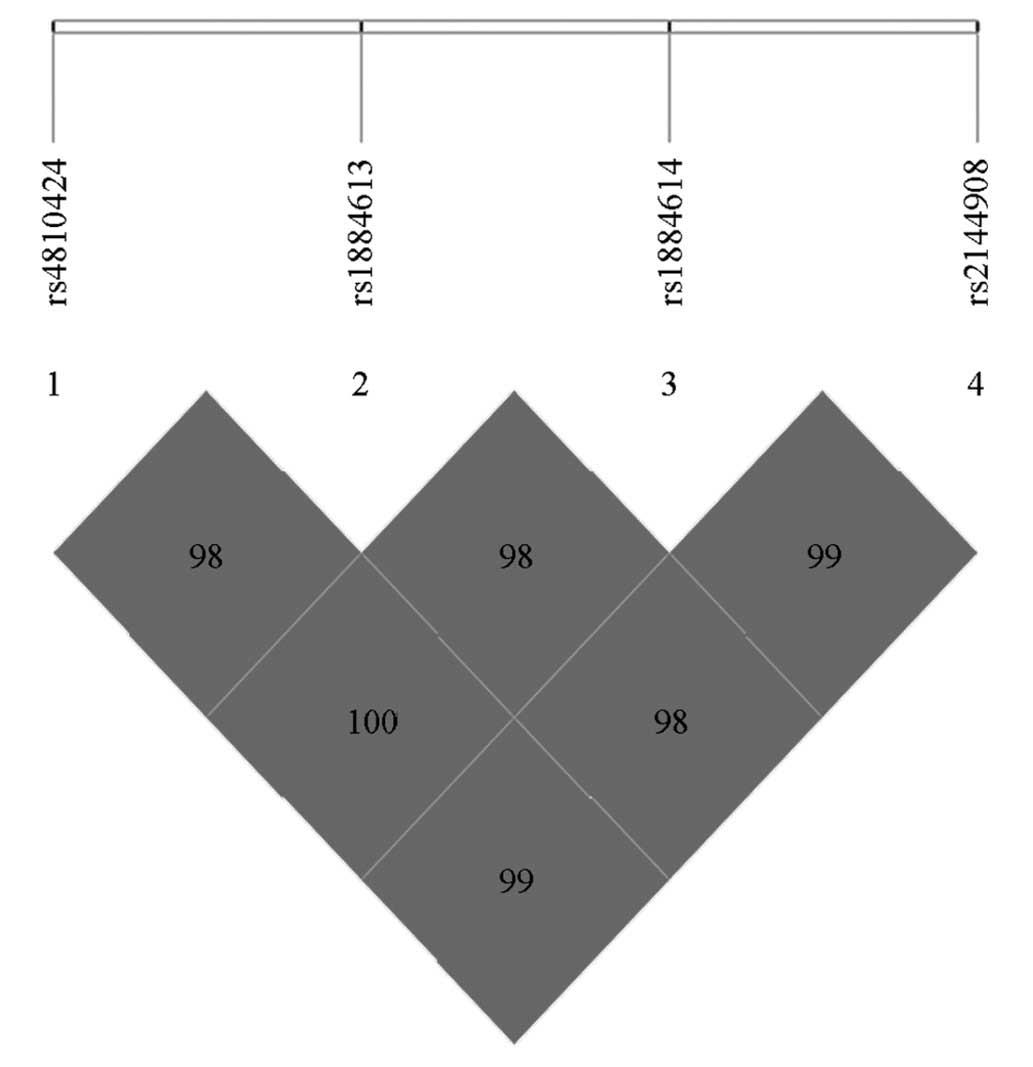
The |D′|values between each two SNPs were calculated
using SHEsis software, to measure the linkage disequilibrium tight
degrees between the loci (Fig. 6).
From the values, it was observed that a marked linkage
disequilibrium existed between the four SNPs, which can build
haploid types. The four SNPs in the HNF-4α gene (rs4810424,
rs1884613, rs1884614 and rs2144908) built three main haploid types:
CGTA, GCCG and CCTA. The frequencies of haplotype CGTA in the type
2 diabetes group and normal control group exhibited a significant
difference (χ2=8.34, P<0.004), while the other
haplotypes revealed no marked differences between the two groups
(Table IV).
 | Table IVCorrelation of the frequencies of
haplotypes in the hepatic nuclear factor-4α gene with type 2
diabetes in Dali Bai individuals. |
Table IV
Correlation of the frequencies of
haplotypes in the hepatic nuclear factor-4α gene with type 2
diabetes in Dali Bai individuals.
| SNPs | Haplotype | T2DM [n (%)] | NC [n (%)] | χ2 | P |
|---|
| rs4810424 +
rs1884613 + rs1884614 + rs2144908 | CGTA | 35.0 (39.7) | 67 (38.5) | 0.11 | 0.74 |
| rs4810424 +
rs1884613 + rs1884614 + rs2144908 | GCCG | 47.0 (53.3) | 107 (61.5) | 1.13 | 0.29 |
| rs4810424 +
rs1884613 + rs1884614 + rs2144908 | CCTA | 4.0 (4.6) | 0 (0.00) | 8.34 | 0.004 |
| rs4810424 +
rs1884613 + rs1884614 + rs2144908 | GGCG | 1.0 (1.2) | 0 (0.00) | NA | NA |
| rs4810424 +
rs1884613 + rs1884614 + rs2144908 | CCCG | 1.0 (1.1) | 0 (0.00) | NA | NA |
Discussion
It is widely reported that the HNF-4α gene is
correlated with type 2 diabetes. In 1998, Hani et al
(18) demonstrated that certain
genetic marker loci in the HNF-4α gene are associated with type 2
diabetes, as determined by a genescan on the French diabetes family
constellations. A missense mutation in HNF-4α +393 makes valine
transform into isoleucine, which is cosegregated with diabetes and
associated with the impaired insulin secretion. It was determined
that the mutation can reduce the transcriptional activity of the
insulin gene in vitro. Subsequently, a study on German
Jewish individuals by Love-Gregory et al (12) also identified that SNPs, including
rs4810424, rsl884613, rsl884614 and rs2144908, in the HNF-4α gene
are closely associated with type 2 diabetes to different extents.
Barroso et al (19) and
Neuman et al (20) also
confirmed the correlation of rs1884613 and/or rs2144908 with type 2
diabetes in a population of Ashkenazi Jews. In Finland, using the
analysis method of the involved siblings, Silander et al
(11) located the susceptibility
genes of type 2 diabetes in chromosome 20 q13.12 to 20 q13.13,
which is the location of the HNF-4α gene. One of the most
significant associated sites with type 2 diabetes is rs2144908,
located near the P2 promoter of HNF-4α gene. However, in Japan,
Hara et al (13) revealed
that the allele and genotype frequencies of two SNPs, rs1884614 and
rs2144908, near the P2 gene promoter of HNF-4α gene was not
associated with type 2 diabetes in a Japanese population. However,
the built haploid type is significantly associated with type 2
diabetes, which is negatively correlated with the distances between
the SNP and the P2 promoter region. However, there are differing
opinions regarding the association of HNF4α gene polymorphisms with
type 2 diabetes. For instance, Tokunaga et al (14) revealed no association between the
haploid type established by rs2144908 and rs1884614, and type 2
diabetes in Japanese individuals. Vaxillaire et al (15) revealed no correlation of the two
SNPs near the P2 promoter with type 2 diabetes in French
individuals. Winckler et al (16) also provided a negative conclusion
in an investigation on the populations in Sweden, Canada and
Finland. In China, no association was observed between rs2144908,
rs1884614 and rs1884613, and type 2 diabetes in Chinese Han
individuals (21–23). Due to the different ethnic
backgrounds and sample sizes, the association of SNPs in the HNF-4α
gene with type 2 diabetes mellitus were ethnically and regionally
different. The different ethnic groups or individuals may cause
different gene types, number and nature of the mutations involved
in diabetes. Therefore, preliminary investigations on different
groups revealed diverse polymorphic loci in the susceptibility
genes.
The inconsistent results from the different areas or
races also reflect the genetic heterogeneity of type 2 diabetes. In
the area 20 q13.12 to 20 q13.13, associated with type 2 diabetes,
there may be genetic heterogeneity and differences caused by the
environmental factors and/or genetic backgrounds. The over
assessment of the high incidence of type 2 diabetes possibly
results from the individuals from the family constellation of type
2 diabetes, however was not ruled out until now. In addition, the
small sample size may also lead to false negative or false positive
results.
In the present study, it was revealed that the
allele and genotype frequencies of four SNPs in the HNF-4α gene
(rs4810424, rsl884613, rsl884614 and rs2144908) presented no
prominent difference between the type 2 diabetes mellitus group and
the healthy control group in the Dali Bai population. However, the
haplotype analysis revealed that there was an associated
disequilibrium between the four SNPs in the HNF-4α gene and the
frequency of the haploid type, CCTA, increased significantly in the
patients with type 2 diabetes mellitus in the Dali Bai population
(P=0.004), suggesting that rs4810424, rsl884613, rsl884614 and
rs2144908 in the HNF-4α gene are not susceptibility loci for type 2
diabetes in the Dali Bai population. However, the existence of the
haploid type, CCTA, built by the four SNPs may increase the risk of
type 2 diabetes in Dali Bai individuals. Haplotype refers to a
collection of specific alleles in a cluster of tightly-linked genes
on a chromosome, which are likely to be inherited together. It has
been reported that haplotype analysis is often more meaningful
compared with a single polymorphic locus analysis (13). According to the results of the
present study, the frequency of haplotype CCTA increased
significantly in the patients with type 2 diabetes mellitus in the
Dali Bai population and it was hypothesized that the synergetic
effect of the four SNPs may change the functional regions of HNF-4α
gene and thereby affect the susceptibility of type 2 diabetes in
the Dali Bai population. Additionally, due to the difficulty of
collecting samples from patients with type 2 diabetes in the Bai
population with three generations of pure Bai line, the small
sample size may lead to certain differences in the results. A
larger sample size would be required in future investigations.
Acknowledgments
The present study was supported by a grant from the
Health Science and Technology Plan of Yunnan Province, China (no.
2011WS0103).
References
|
1
|
Shaw JE, Sicree RA and Zimmet PZ: Global
estimates of the prevalence of diabetes for 2010 and 2030. Diabetes
Res Clin Pract. 87:4–14. 2010. View Article : Google Scholar
|
|
2
|
Tang LL, Liu Q, Bu SZ, Xu LT, Wang QW, Mai
YF and Duan SW: The effect of environmental factors and DNA
methylation on type 2 diabetes mellitus. Yi Chuan. 35:1143–1152.
2013.In Chinese. View Article : Google Scholar
|
|
3
|
Brunetti A, Chiefari E and Foti D: Recent
advances in the molecular genetics of type 2 diabetes mellitus.
World J Diabetes. 5:128–140. 2014.PubMed/NCBI
|
|
4
|
Nammo T, Yamagata K, Tanaka T, Kodama T,
Sladek FM, Fukui K, Katsube F, Sato Y, Miyagawa J and Shimomura I:
Expression of HNF-4alpha (MODY1), HNF-1beta (MODY5), and HNF-1alpha
(MODY3) proteins in the developing mouse pancreas. Gene Expr
Patterns. 8:96–106. 2008. View Article : Google Scholar
|
|
5
|
Babeu JP and Boudreau F: Hepatocyte
nuclear factor 4-alpha involvement in liver and intestinal
inflammatory networks. World J Gastroenterol. 20:22–30. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gupta RK, Vatamaniuk MZ, Lee CS, Flaschen
RC, Fulmer JT, Matschinsky FM, Duncan SA and Kaestner KH: The MODY1
gene HNF-4alpha regulates selected genes involved in insulin
secretion. J Clin Invest. 115:1006–1015. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yamagata K, Furuta H, Oda N, Kaisaki PJ,
Menzel S, Cox NJ, Fajans SS, Signorini S, Stoffel M and Bell GI:
Mutations in the hepatocyte nuclear factor-4α gene in
maturity-onset diabetes of the young (MODY1). Nature. 384:458–460.
1996. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Johansson S, Raeder H, Eide SA, Midthjell
K, Hveem K, Søvik O, Molven A and Njølstad PR: Studies in 3,523
Norwegians and meta-analysis in 11,571 subjects indicate that
variants in the hepatocyte nuclear factor 4 alpha (HNF4A) P2 region
are associated with type 2 diabetes in Scandinavians. Diabetes.
56:3112–3117. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Cho YS, Chen CH, Hu C, Long J, Ong RT, Sim
X, Takeuchi F, Wu Y, Go MJ, Yamauchi T, et al MuTHER Consortium:
Meta-analysis of genome-wide association studies identifies eight
new loci for type 2 diabetes in east Asians. Nat Genet. 44:67–72.
2011. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Jafar-Mohammadi B1, Groves CJ, Gjesing AP,
Herrera BM, Winckler W, Stringham HM, Morris AP, Lauritzen T, Doney
AS, Morris AD, et al DIAGRAM Consortium: A role for coding
functional variants in HNF4A in type 2 diabetes susceptibility.
Diabetologia. 54:111–119. 2011. View Article : Google Scholar :
|
|
11
|
Silander K, Mohlke KL, Scott LJ, Peck EC,
Hollstein P, Skol AD, Jackson AU, Deloukas P, Hunt S, Stavrides G,
et al: Genetic variation near the hepatocyte nuclear factor-4α gene
predicts susceptibility to type 2 diabetes. Diabetes. 53:1141–1149.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Love-Gregory LD, Wasson J, Ma J, Jin CH,
Glaser B, Suarez BK and Permutt MA: A common polymorphism in the
upstream promoter region of the hepatocyte nuclear factor-4α gene
on chromosome 20q is associated with type 2 diabetes and appears to
contribute to the evidence for linkage in an ashkenazi jewish
population. Diabetes. 53:1134–1140. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hara K, Horikoshi M, Kitazato H, Ito C,
Noda M, Ohashi J, Froguel P, Tokunaga K, Tobe K, Nagai R, et al:
Hepatocyte nuclear factor-4alpha P2 promoter haplotypes are
associated with type 2 diabetes in the Japanese population.
Diabetes. 55:1260–1264. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tokunaga A, Horikawa Y, Fukuda-Akita E,
Okita K, Iwahashi H, Shimomura I, Takeda J and Yamagata K: A common
P2 promoter polymorphism of the hepatocyte nuclear factor-4α gene
is associated with insulin secretion in non-obese Japanese with
type 2 diabetes. Endocr J. 55:999–1004. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Vaxillaire M, Dina C, Lobbens S, Dechaume
A, Vasseur-Delannoy V, Helbecque N, Charpentier G and Froguel P:
Effect of common polymorphisms in the HNF4alpha promoter on
susceptibility to type 2 diabetes in the French Caucasian
population. Diabetologia. 48:440–444. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Winckler W, Graham RR, de Bakker PI, Sun
M, Almgren P, Tuomi T, Gaudet D, Hudson TJ, Ardlie KG, Daly MJ, et
al: Association testing of variants in the hepatocyte nuclear
factor 4alpha gene with risk of type 2 diabetes in 7,883 people.
Diabetes. 54:886–892. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
World Health Organization: Definition,
Diagnosis and Classification of Diabetes Mellitus and its
Complications: Report of a WHO Consultation. Part 1: Diagnosis and
Classification of Diabetes Mellitus. World Health Organization;
Geneva: 1999
|
|
18
|
Hani EH, Suaud L, Boutin P, Chèvre JC,
Durand E, Philippi A, Demenais F, Vionnet N, Furuta H, Velho G, et
al: A missense mutation in hepatocyte nuclear factor-4 alpha,
resulting in a reduced transactivation activity, in human
late-onset non-insulin-dependent diabetes mellitus. J Clin Invest.
101:521–526. 1998. View
Article : Google Scholar : PubMed/NCBI
|
|
19
|
Barroso I, Luan J, Wheeler E, Whittaker P,
Wasson J, Zeggini E, Weedon MN, Hunt S, Venkatesh R, Frayling TM,
et al: Population-specific risk of type 2 diabetes conferred by
HNF4A P2 promoter variants. Diabetes. 57:3161–3165. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Neuman RJ, Wasson J, Atzmon G, Wainstein
J, Yerushalmi Y, Cohen J, Barzilai N, Blech I, Glaser B and Permutt
MA: Gene-gene interactions lead to higher risk for development of
type 2 diabetes in an Ashkenazi Jewish population. PLoS One.
5:e99032010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang F, Han XY, Ren Q, Zhang XY, Han LC,
Luo YY, Zhou XH and Ji LN: Effect of genetic variants in KCNJ11,
ABCC8, PPARG and HNF4A loci on the susceptibility of type 2
diabetes in Chinese Han population. Chin Med J (Engl).
122:2477–2482. 2009.
|
|
22
|
Chen Z, Zhang D, Liu Y, Zhou D, Zhao T,
Yang Y, He L and Xu H: Variants in hepatocyte nuclear factor 4α
gene promoter region and type 2 diabetes risk in Chinese. Exp Biol
Med (Maywood). 235:857–861. 2010. View Article : Google Scholar
|
|
23
|
Wang C, Chen S, Zhang T, Chen Z, Liu S,
Peng X, Ma J, Zhong X, Yan Y, Tang L, et al: Prediabetes is
associated with HNF-4α P2 promoter polymorphism rs1884613: A
case-control study in Han Chinese population and an updated
meta-analysis. Dis Markers. 2014:2317362014. View Article : Google Scholar
|