Introduction
Gastric cancer is a common type of gastrointestinal
cancer worldwide (1). In China,
the incidence of gastric cancer is the highest of the digestive
system neoplasms and second for all malignant tumors (2). Recent studies have observed that
there are ~500,000 diagnoses of gastric cancer in China every year,
accounting for ~50% of diagnoses worldwide. Furthermore, stomach
cancer results in 10% of cancer-associated mortalities (3). Investigations into the association
between microRNA (miR) and stomach cancer have increased. miRs are
a class of non-coding RNA 18–21 nt in length. They are highly
expressed in eukaryotic cells, and contribute to the regulation of
cell growth, development and apoptosis (4,5).
Numerous studies have shown that the occurrence and progression of
tumors is accompanied by an abnormal change in miR expression
levels (6,7). A number of studies have demonstrated
increased expression levels of miR-21 in various solid tumors, for
example, Volinia et al (8)
observed that miR-21 was upregulated in breast and lung cancer, and
was associated with a poor prognosis. However, the association
between miR-21 expression and gastric cancer proliferation,
invasion and migration has not been widely investigated, and the
underlying mechanism of the potential target protein miR-21 remains
to be elucidated. The present study aimed to validate the
association between miR-21 and the clinical features of patients
with gastric cancer, as well as the proliferation, invasion and
migration of gastric cancer cells by analyzing samples of gastric
cancer tissues and the results of in vitro cellular
experiments. Bioinformatics and dual luciferase report analysis in
the SGC-7901 gastric cancer cell line was performed in the current
study and demonstrated that Noxa, a member of the B-cell lymphoma 2
(Bcl-2) family with pro-apoptotic effects (9–11) is
the target gene of miR-21.
Materials and methods
Clinical data of patients
Paraffin-embedded tissue samples were collected from
40 patients, who had undergone radical resection for gastric cancer
in the Fifth Affiliated Hospital of Zhengzhou University
(Zhengzhou, China) between January 2012 and May 2014. Patients
included 22 males and 18 females, with a median age of 60 years
(range, 43–76 years). All patients were diagnosed by endoscopy and
pathology. In eight cases the tumor size was <3 cm, in 10 cases
the tumor size was 3–5 cm, and in 22 cases the tumor size was ≥5
cm; 24 cases were intestinal-type and 16 cases were diffuse-type.
There were 14 cases with lymph node metastasis, and 26 cases
without lymph node metastasis. TNM staging was conducted (12) and 7 cases were stage I, 9 cases
were stage II, 18 cases were stage III and 6 cases were stage IV
(Table I). The patients had not
received preoperative chemotherapy or radiotherapy. The present
study was approved by the ethics committee of the Fifth Affiliated
Hospital of Zhengzhou University (Zhengzhou, China).
 | Table IAssociation between miR-21 expression
and pathological characteristics of patients with gastric
cancer. |
Table I
Association between miR-21 expression
and pathological characteristics of patients with gastric
cancer.
| Clinical feature | Cases | Expression level of
miR-21 (2−ΔΔCq)
| Statistical
value | P-value |
|---|
| <4.23 | ≥4.23 |
|---|
| Gender | | | | 0.017 | P>0.05 |
| Male | 22 | 9 | 13 | | |
| Female | 18 | 7 | 11 | | |
| Age (years) | | | | 3.300 | P>0.05 |
| <60 | 22 | 6 | 16 | | |
| ≥60 | 18 | 10 | 8 | | |
| TNM stage | | | | 5.625 | P<0.05 |
| Low (Ⅰ and
II) | 16 | 10 | 6 | | |
| High (III and
Ⅳ) | 24 | 6 | 18 | | |
| Lymph node
metastasis | | | | 8.864 | P<0.05 |
| Yes | 14 | 10 | 4 | | |
| No | 26 | 6 | 20 | | |
| Histological
type | | | | 1.111 | P>0.05 |
| Intestinal | 24 | 8 | 16 | | |
| Diffuse | 16 | 8 | 8 | | |
| Tumor size
(cm) | | | | 6.077 | P<0.05 |
| <5 | 18 | 11 | 7 | | |
| ≥5 | 22 | 5 | 17 | | |
Detection of miR-21 in tissue
samples
The miR was extracted and purified from cancer
tissues and matched adjacent non-tumor tissues of the 40 patients
using a paraffin-embedded tissue microRNA rapid extraction kit
(Beijing Bioteke Biotechnology Co., Ltd., Beijing, China) according
to the manufacturer's protocols. In brief, tissue samples were
treated with dimethylbenzene to remove the paraffin and then
guanidinium isothiocyanate was added for tissue lysis. Lysates were
treated with chloroform and centrifuged at 12,000 × g for 15 min
according to the manufacturer's instructions of the protein extract
kit (Thermo Fisher Scientific, Inc., Waltham, MA, USA). miRNA
products were purified using a silicon substrate membrane
adsorption column from the extraction kit. Reverse transcription
(RT) of miR into cDNA was performed using a One Step PrimeScript
miRNA cDNA Synthesis kit (Takara Biotechnology, Co., Ltd., Dalian,
China) according to the manufacturer's protocols. For each sample,
2 µg miR was reverse transcribed in the reaction system,
including miR reaction buffer mix (containing oligodt and RT
primers miRNA) and PrimeScript® RT enzyme mix
(containing poly(A) polymerase and reverse transcriptase). For
setting the RT− control, the same system was used,
however, water was used to replace miR. The reaction conditions
were as follows: 60 min poly(A) polymerase reaction and reverse
transcription at 37°C and 5 sec at 85°C for inactivating the
enzymes. The cDNA was amplified by RT quantitative polymerase chain
reaction (qPCR) with the SYBR® Premix Ex Taq™ II kit
(Takara Biotechnology, Co., Ltd.) according to the manufacturer's
protocols. The primers were as follows: Sense,
5′-GCCGCTAGCTTATCAGACTGATGT-3′ for miR-21; and sense,
5′-CTCGCTTCGGCAGCACA-3′ for U6 snRNA, which was used to normalize
the data. The antisense primer, Uni-miR qPCR, was obtained from the
One Step PrimeScript miRNA cDNA Synthesis kit. The relative
expression level of miR-21 was expressed by 2−ΔΔCq (for
tumor tissue/non-tumor tissues), where 2−ΔΔCq=Cq
(target)-Cq (U6 snRNA) (13).
Detection of Noxa expression levels in
tissue samples
The total RNA of tumor and non-tumor tissues was
extracted using TRIzol® (Thermo Fisher Scientific,
Inc.), and RT of miR into cDNA was performed using the GoScript
Reverse Transcription kit [Promega (Beijing) Biotech Co., Ltd.,
Beijing, China] according to the manufacturer's protocols.
Fluorescent RT-qPCR was implemented using a GoTaq® qPCR
kit [Promega (Beijing) Biotech Co., Ltd.] according to the
manufacturer's protocols. Primers used were as follows: Sense,
5′-TTCGTGTTCAGCTCGCGTCC-3′ and antisense,
5′-CTCGGTGTAGCCTTCTTGCC-3′ for Noxa; and sense,
5′-ACCACAGTCCATGCCATCAC-3′ and antisense,
5′-TCCACCACCCTGTTGCTGTA-3′ for GAPDH, which was used to normalize
the data. The relative expression level of Noxa was expressed by
2−ΔΔCq (for tumor tissue / non-tumor tissue).
Total protein of the tissue sample was extracted by
radioimmunoprecipitation assay (RIPA) lysis buffer (Thermo Fisher
Scientific, Inc.) containing 1% protease inhibitor cocktail (500
µl/well; Thermo Fisher Scientific, Inc.). Electrophoresis
was performed with 12% polyacrylamide gel (Applygen Technologies
Inc., Beijing, China) at 100 V for 1.5 h and for each sample, 45
µg protein was loaded onto each lane. Following
electrophoresis, the protein bands were transferred to a
polyvinylidene fluoride membrane (Applygen Technologies, Inc.) with
a 200-mA constant current for 1.5 h. The membranes were blocked for
1 h with 5% skimmed milk powder at room temperature prior to
incubation with mouse monoclonal anti-human Noxa antibody (1:500;
Santa Cruz Biotechnology, Inc., Dallas, TX, USA; cat. no.
sc-398873) and mouse monoclonal anti-human β-actin antibody
(1:1,000; Santa Cruz Biotechnology, Inc.; cat. no. sc-47778) at 4°C
overnight. The membranes were washed three times with
phosphate-buffered saline with Tween-20 (Applygen Technologies,
Inc.) to remove excess antibodies, and the horseradish
peroxidase-labeled goat anti-mouse IgG (1:500; Santa Cruz
Biotechnology, Inc; cat. no. sc-2005) secondary antibody was added
and incubated for 2 h at room temperature. The membranes were
washed to remove excess antibodies and color development of bands
was visualized using an enhanced chemiluminescence color
development kit (Applygen Technologies, Inc.), and the Noxa protein
band intensity was analyzed using the BandScan system (Tanon 5200
Image Analysis System; Tanon Science & Technology Co., Ltd.,
Shanghai, China).
Culture of the SGC-7901 gastric cancer
cell line
The SGC-7901 human gastric cell line was purchased
from the cell bank of the Shanghai Institute of Biochemistry and
Cell Biology (Chinese Academy of Sciences, Shanghai, China).
Dulbecco's modified Eagle's medium (DMEM; Thermo Fisher Scientific,
Inc.) containing 10% fetal bovine serum (FBS; Thermo Fisher
Scientific, Inc.) was used for cell culture and 0.25% trypsin
(Thermo Fisher Scientific, Inc.) was used for cell digestion. Cells
were cultured at 37°C in 5% CO2 for 24–48 h.
Upregulation of miR-21
miR-21 mimics were designed and synthesized by
Shanghai GenePharma Co., Ltd. (Shanghai, China). SGC-7901 cells in
the logarithmic growth phase were transfected with the miR-21 mimic
and the miR-negative control (NC) using a Lipofectamine™ 2000
transfection kit (Thermo Fisher Scientific, Inc.). The transfection
was conducted according to the manufacturer's protocol.
Overexpression of miR-21 was verified using RT-qPCR 48 h following
transfection.
Effect of miR-21 upregulation on SGC-7901
cell proliferation and the cell cycle
The non-transfected (control), miR-21
mimic-transfected and miR-NC-transfected SGC-7901 cells were
maintained in 96-well plates at a density of 1×104/well
for 6, 12, 24 or 48 h. MTT reagent (10 µl) was added to each
well, and after 4 h, DMSO (100 µl) was added to each well.
The optical density (OD) values at 490 nm were determined using a
microplate reader (UV/Vis Absorbance Spectra; BMG Labtech, Ltd.,
Cary, NC, USA). For cell cycle analysis, transfected and
non-transfected cells were fixed in cold 70% ethanol and stained
with 50 µg/ml propidium iodide (Sigma-Aldrich, St. Louis,
MO, USA) and incubated for 30 min at room temperature. Analysis was
performed using a BD FACSCalibur flow cytometer (Becton, Dickinson
and Company, San Jose, CA, USA) within 15 min after incubation.
CellQuest software (version 5.1; Becton, Dickinson and Company) was
used to analyze the cell cycle and apoptosis rate.
Effect of miR-21 upregulation on SGC-7901
cell migration and invasion
The non-transfected (control), miR-21
mimic-transfected and miR-NC-transfected SGC-7901 cells were
cultured in an uncoated Transwell chamber at a density of
1×105/well in the upper layer (membrane pore size, 8
µm) to detect the migration of cells. The above-mentioned
cells were also cultivated in a Matrigel-coated Transwell chamber
(Corning Inc., Corning, NY, USA) in the upper layer (membrane pore
size, 8 µm) to detect the invasive ability of the cells. The
serum-free DMEM medium was used for the upper chamber and the
medium containing 10% FBS was placed in the lower chamber.
Following culture for 6 h, the cells that had remained in the upper
chamber were removed, and the number of cells that had migrated or
invaded the lower layer was estimated using Giemsa (Applygen
Technologies, Inc.) staining.
Prediction and verification of miR-21
targets
The target gene of miR-21 was predicted using the
bioinformatics prediction software, PicTar (http://pictar.mdc-berlin.de/) and TargetScan5.0
(Whitehead Institute for Biomedical Research, Cambridge, MA, USA).
Following integration of the results from the software, the
candidate target genes were further confirmed by the dual
luciferase reporting system. Shanghai Sangong Pharmaceutical Co.,
Ltd. (Shanghai, China) designed and constructed the luciferase
reporter plasmid containing wild-type Noxa 3′-untranslated region
(UTR) and mutant Noxa 3′-UTR, designated pMIR-Noxa-wild and
pMIR-Noxa-mut, respectively. The SGC-7901 cells were divided into
four groups as follows: miR-21 mimic + pMIR-Noxa-wild; miR-NC +
pMIR-Noxa-wild; miR-21 mimic + pMIR-Noxa-mut; and miR-NC +
pMIR-Noxa-wild. Following transfection, cells were cultivated in
96-well plates at a density of 1×104/well and cultured
for 48 h, prior to the addition of 100 µl lysis buffer
(Applygen Technologies, Inc.) to each well. The supernatant was
collected following lysis, and 10 µl of supernatant was
added to the 40 µl firefly luciferase substrate and mixed
evenly for 15 sec prior to determining the fluorescence intensity.
The remaining supernatant was added to 40 µl Renilla
luciferase substrate, and mixed evenly before determining the
fluorescence intensity. The ratio of firefly to Renilla
luciferase intensity was calculated to demonstrate the effect of
miR-21 on the expression level of Noxa mRNA.
Effect of miR-21 overexpression on Noxa
expression levels
The total RNA of non-transfected, miR-21
mimic-transfected and miR-NC-transfected SGC-7901 cells was
extracted using TRIzol®. The RT of miR into cDNA was
performed using the GoScript Reverse Transcription kit [Promega
(Beijing) Biotech Co., Ltd.] according to the manufacturer's
protocols. Fluorescent RT-qPCR was implemented using a
GoTaq® qPCR kit according to the manufacturer's
protocols. The sequence of the primers for Noxa was as described
above. The total protein in the cells was extracted using RIPA
lysis buffer (500 µl/well) containing 1% protease inhibitor
cocktail. The effect of miR-21 upregulation on Noxa protein
expression levels was detected by western blotting, following the
above-mentioned procedure.
Statistical analysis
Statistical analysis was performed using SPSS 13.0
statistical software (SPSS, Inc., Chicago, IL, USA). The normality
data are expressed as means ± standard deviation and the non-normal
data are expressed as medians. Data were compared between two or
multiple groups using Student's t-test and one-way analysis of
variance, respectively. The non-parametric test was performed by
Mann-Whitney rank-sum test and χ2 test. P<0.05 was
considered to indicate a statistically significant difference.
Results
Increased expression levels of miR-21 in
gastric cancer tissue are associated with the clinical features of
gastric cancer
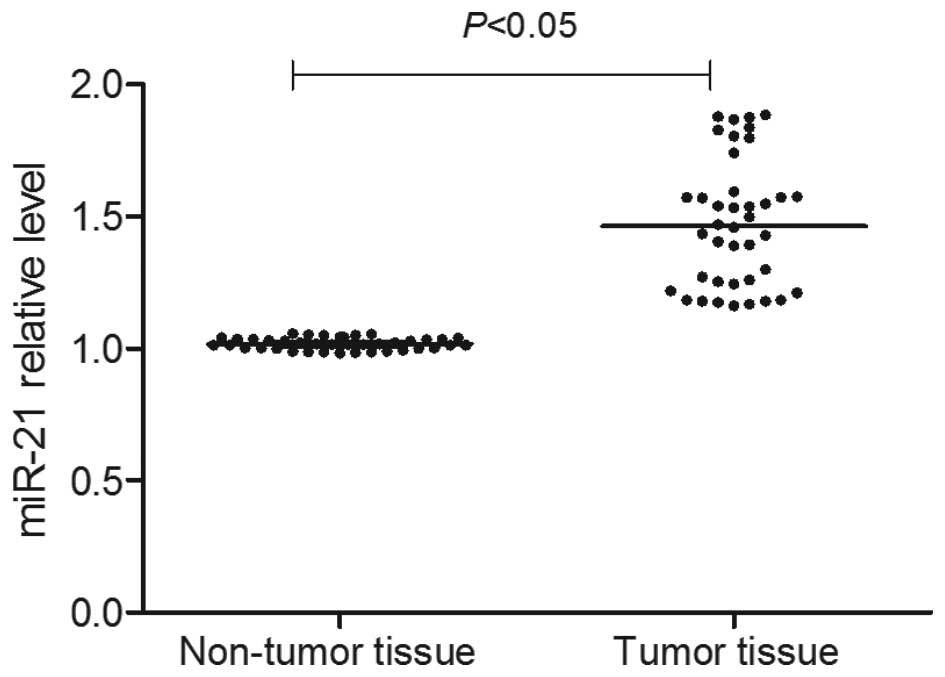
As presented in Fig.
1, RT-qPCR detection demonstrated that the miR-21 expression
level in the cancer tissue was significantly increased (P<0.05).
Using the median of the miR-21 expression levels
(2−ΔΔCq) in gastric cancer tissue as the limit, all
patients with gastric cancer were divided into an miR-21 high
expression group [2−ΔΔCq (miR-21)≥4.23] and an miR-21
low expression group [2−ΔΔCq (miR-21)<4.23]. Analysis
of the association between the clinical features of patients
(gender, age, tumor size, histological type, lymph node metastasis
and TNM staging), and the level of miR-21 expression (Table I) demonstrated that the expression
level of miR-21 was not associated with the patient's age, gender,
and histological type (P>0.05); however was associated with the
tumor size, TNM stage and presence of lymph node metastasis
(P<0.05). Univariate analysis demonstrated that increased
expression levels of miR-21 were associated with larger tumor size
[diameter, >5 cm; odds ratio (OR), 5.34; 95% confidence interval
(CI), 1.35–21.15; P<0.05], high TNM stage (stage III and IV; OR,
5.00; 95% CI, 1.27–19.68; P<0.05), and the emergence of lymph
node metastasis (OR, 8.33; 95% CI, 1.90–6.44; P<0.05).
Expression levels of Noxa in gastric
cancer tissue were decreased and negatively correlated with the
expression level of miR-21
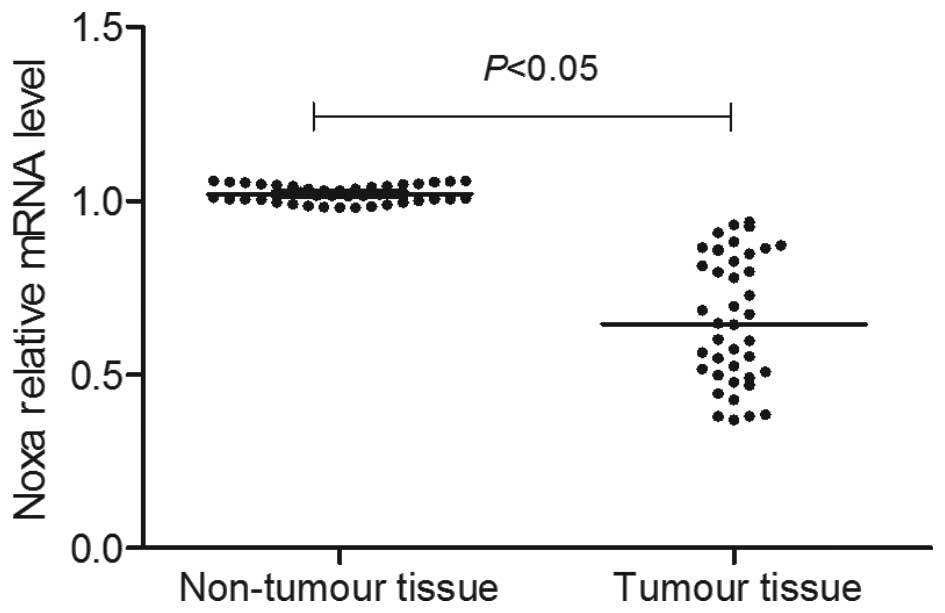
As presented in Fig.
2, RT-qPCR detection demonstrated that the mRNA level of Noxa
in tumor tissue was significantly lower than that of the adjacent
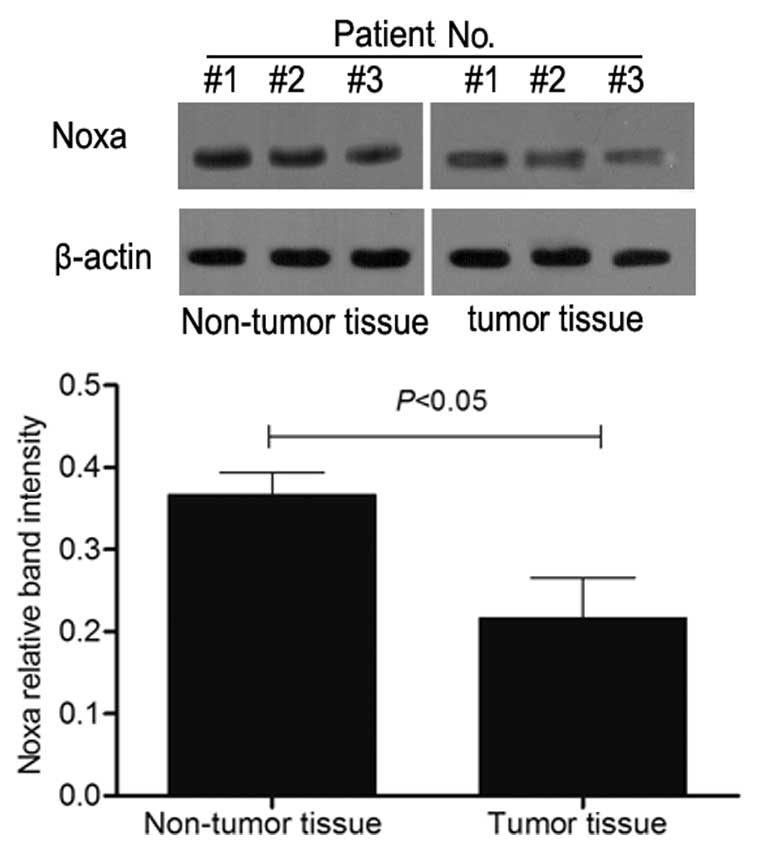
non-tumor tissue samples (P<0.05). Consistent with the RT-qPCR
results, the western blot analysis indicated that the Noxa protein
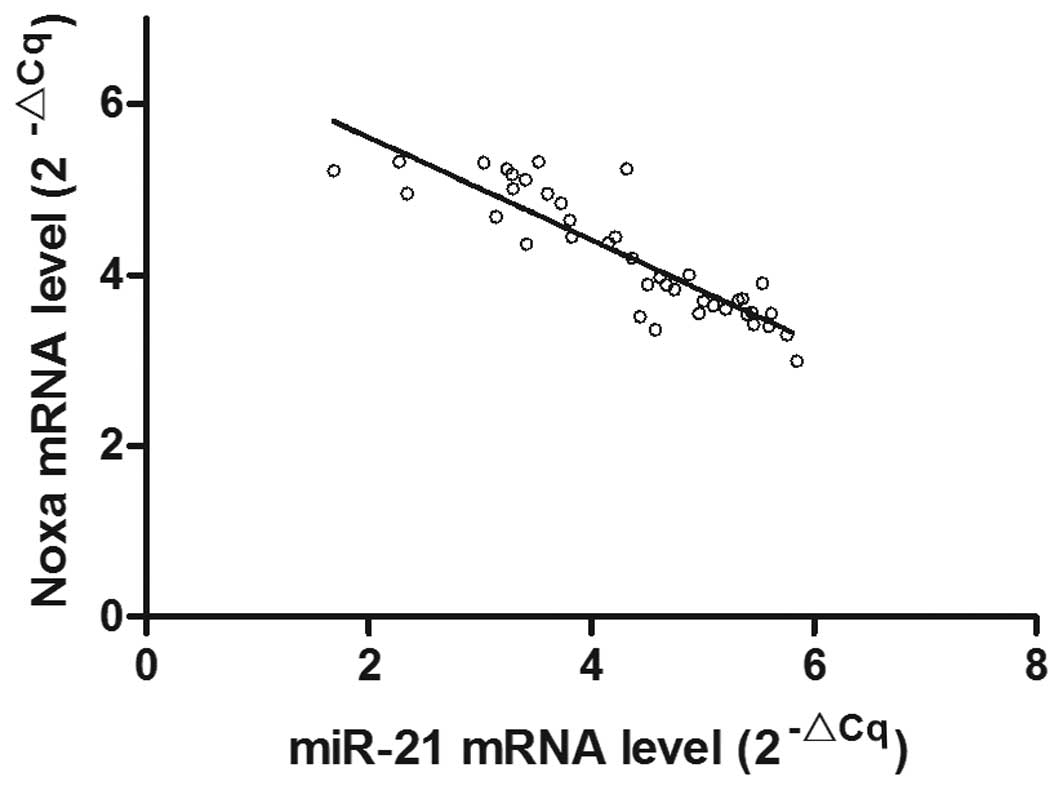
expression level in the tumor tissue was decreased (Fig. 3). The correlation analysis
(Fig. 4) demonstrated that the
mRNA expression level of Noxa in the gastric cancer tissue samples
was inversely proportional to the level of miR-21 mRNA expression
(r=−0.782; P<0.05).
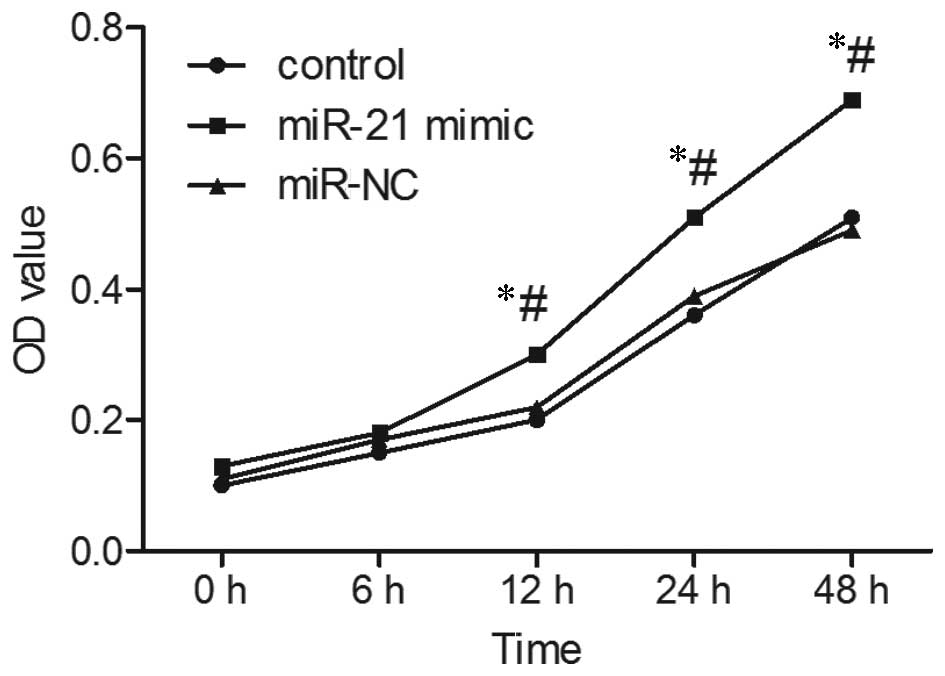
Upregulation of miR-21 promotes growth of
SGC-7901 cells
As presented in Fig.
5, MTT analysis indicated that following upregulation of
miR-21, the rate of proliferation of SGC-7901 cells was
significantly increased. Following culture for 12 and 48 h, the OD
value of SGC-7901 cells following upregulation of miR-21 was
significantly higher than that of the non-transfected (control) and
miR-NC-transfected SGC-7901 cells (P<0.05). Cell cycle analysis
revealed that following transfection of the miR-21 mimic, the ratio
of G0/G1 of SGC-7901 cells decreased, while
the proportion of cells in the S phase increased, indicating active
DNA synthesis in the cells (Fig.
6).
Increased expression levels of miR-21
promoted the migration and invasion of SGC-7901 cells
As presented in Fig.
7, the Transwell chamber culture analysis demonstrated that,
compared with the non-transfected SGC-7901 cells and SGC-7901 cells
transfected with miR-NC, the SGC-7901 cells transfected with miR-21
mimic exhibited marked migration and invasion.
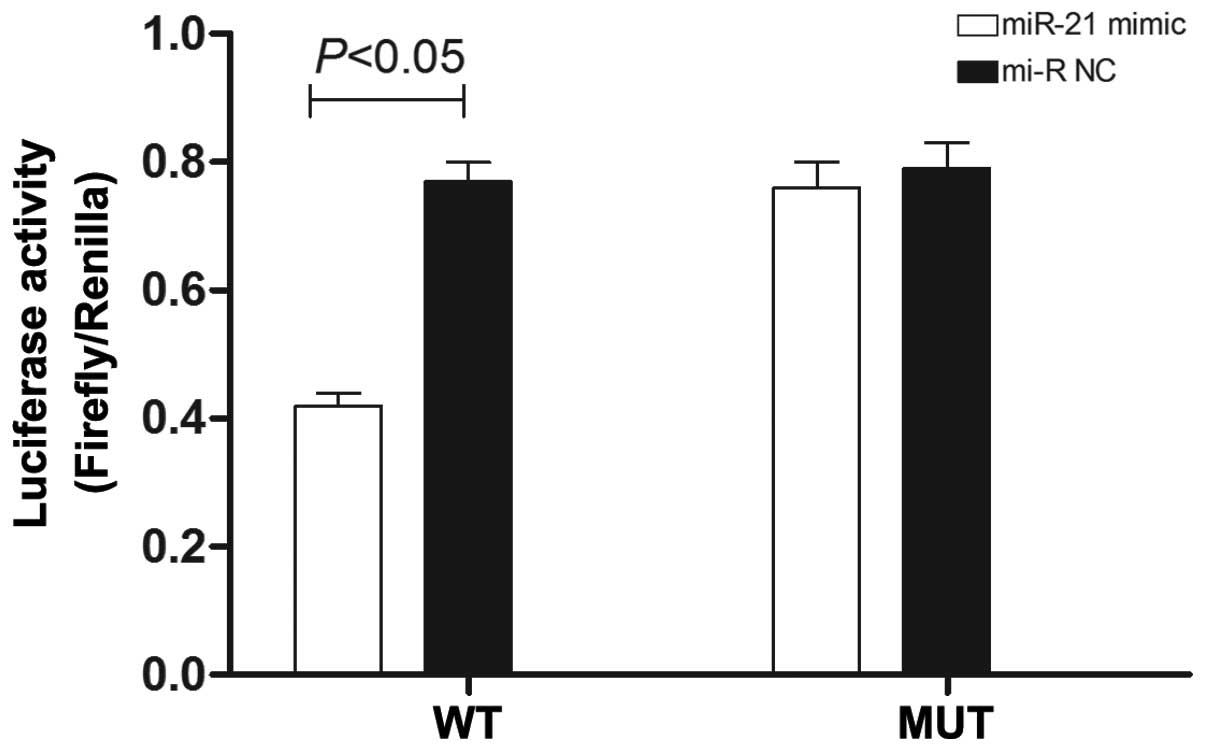
Noxa is a potential target of miR-21
The analysis results using PicTar and TargetScan5.0
software indicated that the microRNA gene binding free energy of
the Noxa gene binding with miR-21 was lower than other potential
miR-21-targeted mRNAs, indicating that miR-21 can easily bind with
Noxa, and thus Noxa may be a target of miR-21. The complementary
base pairing existed between seven bases of miR-21 and the 3′-UTR.
Thus, the Noxa gene may be a potential target gene of miR-21. The
dual luciferase report analysis (Fig.
8) demonstrated that the fluorescence intensity of SGC-7901
cells transfected with miR-21 mimic + pMIR-Noxa-wild was
significantly lower than that of the cells transfected with miR-NC
+ pMIR-Noxa-wild (P<0.05). The fluorescence value of SGC-7901
cells transfected with miR-21 mimic + pMIR-Noxa-mut was not
decreased significantly compared with the cells transfected with
miR-NC + pMIR-Noxa-mut, suggesting that miR-21 may exert an
inhibitory effect by functioning at the 3′-UTR region of Noxa.
RT-qPCR and western blotting (Figs.
9 and 10) confirmed that Noxa
may be a target gene of miR-21.
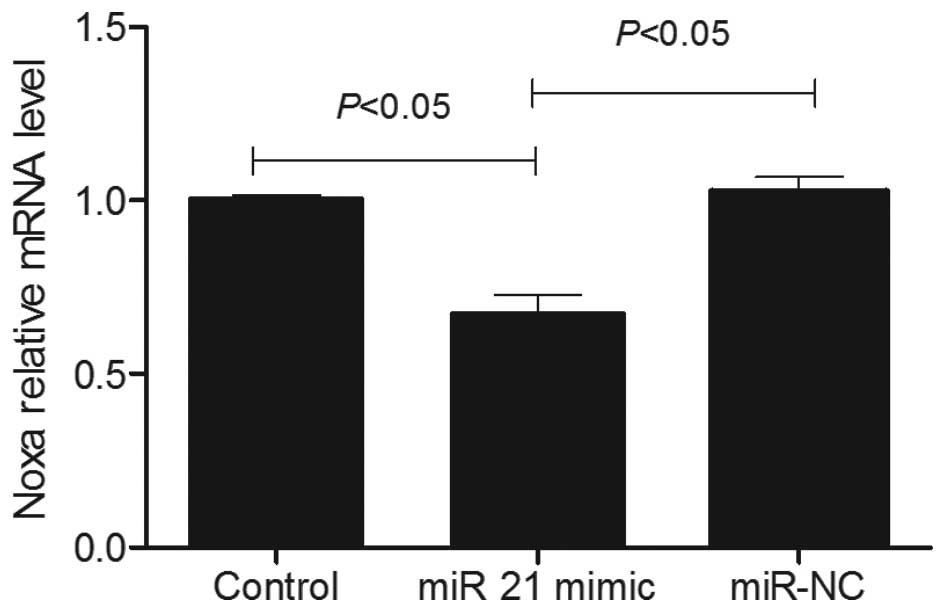
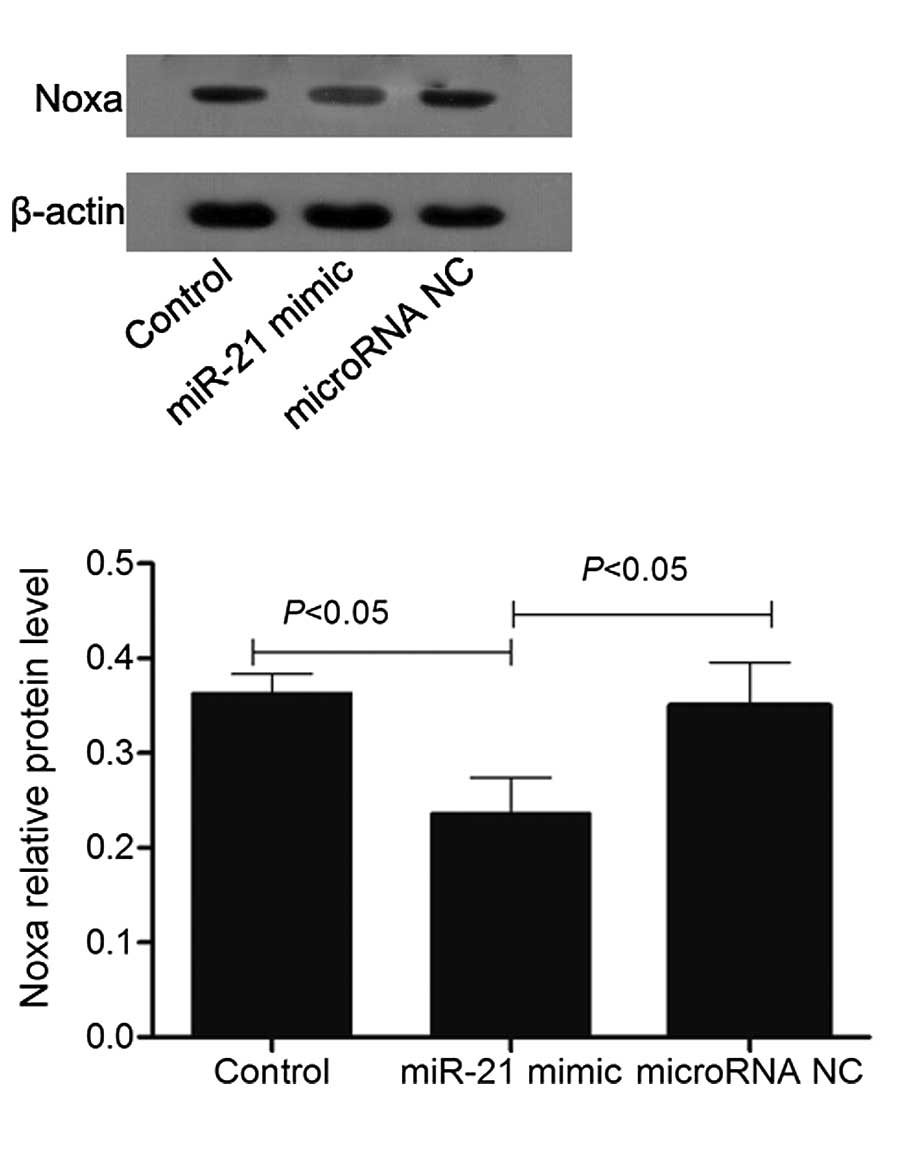
Overexpression of miR-21 reduced the
expression levels of Noxa
As presented in Fig.
9, RT-qPCR analysis demonstrated that the mRNA level of Noxa in
SGC-7901 cells transfected with the miR-21 mimic was decreased
compared with the non-transfected SGC-7901 cells and SGC-7901 cells
transfected with miR NC. It was decreased by 0.63±0.03-fold vs. the
non-transfected SGC-7901 cells (P<0.05). The results of the
western blot analysis were consistent with those of the RT-qPCR,
demonstrating that an increased level of miR-21 augments the
expression level of Noxa protein in SGC-7901 cells (Fig. 10).
Discussion
miRs have wide-ranging applications as tumor
biomarkers. However, the association between miRs and
tumorigenesis, metastasis and infiltration, as well as the
underlying mechanisms, requires further investigation. miR-21 is
present in a variety of solid tumors. Numerous studies have
demonstrated increased expression levels of miR-21 in breast,
hepatocellular, lung, pancreatic and ovarian cancer, and glioma
(14,15). Furthermore, increased expression
levels of miR-21 are associated with poor patient prognosis
(16–19). In the present study, the change of
miR-21 expression level in patients with gastric cancer was
analyzed. The tissue sample analysis demonstrated that the
expression level of miR-21 in the cancer tissue samples was
significantly increased compared with that of non-tumor tissue
samples. Further analysis also indicated that the expression level
of miR-21 was associated with the clinical features of patients
with gastric cancer; for example, the miR-21 expression level of
patients at an advanced TNM stage and patients with lymph node
metastasis was increased. Following upregulation of miR-21,
SGC-7901 proliferation was observed to accelerate. In addition, the
cell cycle analysis indicated an increased proportion of cells in S
phase with greater invasion and migration capability.
Noxa is a member of the Bcl-2 family, which is
located in the mitochondrial membrane and exerts pro-apoptotic
effects (20,21). Its mechanism of pro-apoptosis is
associated with the mitochondrial cytochrome c signaling
pathway. Noxa increases the permeability of the mitochondrial
membrane, promoting the release of cytochrome c (9,10).
It is a downstream gene of p53 and when cells are damaged following
stimulation, p53 binds with the upstream promoter sequence of Noxa
to enhance expression levels. Thus, the pro-apoptotic effects of
Noxa are dependent on p53 (22–24).
In tumor cells, Noxa expression levels decrease, which may affect
tumor cell apoptosis and reduce the effect of therapeutic agents
(25). For example, Kuwahara et
al (26) found that decreased
Noxa expression levels may result in decreased sensitivity to
chemotherapeutic agents in malignant rhabdoid tumor. Ehrhardt et
al (27) observed that
following p53 and Noxa gene knockout, apoptosis of acute
lymphoblastic leukemia, induced by conventional cytotoxic
therapeutic agents combined with betulinic acid, decreased.
Consistent with these previous findings, decreased expression
levels of Noxa were observed in gastric cancer tissue samples in
the present study. In addition, Noxa expression levels in gastric
cancer patients were observed to be negatively correlated with the
miR-21 expression level. The bioinformatics software and dual
luciferase reporter enzymes indicated that Noxa is the target gene
of miR-21 and upregulation of miR-21 in SGC-7901 cells reduced the
Noxa expression levels. Therefore, miR-21 and Noxa may serve as
targets for the treatment of gastric cancer. However, the present
study did not clarify the effect of variations in the expression
level of Noxa on the gastric cancer cells, which requires further
investigation.
In conclusion, miR-21 affects the proliferation,
invasion and migration of gastric cancer cells via Noxa. Increased
expression levels of miR-21 and decreased expression levels of Noxa
decreases apoptosis of tumor cells and results in a poor prognosis
for gastric cancer patients.
References
|
1
|
Ferro A, Peleteiro B, Malvezzi M, Bosetti
C, Bertuccio P, Levi F, Negri E, La Vecchia C and Lunet N:
Worldwide trends in gastric cancer mortality (1980–2011), with
predictions to 2015, and incidence by subtype. Eur J Cancer.
50:1330–1344. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ding YB, Xia TS, Wu JD, Chen GY, Wang S
and Xia JG: Surgical outcomes for gastric cancer of a single
institute in southeast China. Am J Surg. 203:217–221. 2012.
View Article : Google Scholar
|
|
3
|
Wang R and Chen XZ: High mortality from
hepatic, gastric and esophageal cancers in mainland China: 40 years
of experience and development. Clin Res Hepatol Gastroenterol.
38:751–756. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bhattacharyya M, Nath J and Bandyopadhyay
S: MicroRNA signatures highlight new breast cancer subtypes. Gene.
556:192–198. 2015. View Article : Google Scholar
|
|
5
|
Rane JK, Scaravilli M, Ylipää A, Pellacani
D, Mann VM, Simms MS, Nykter M, Collins AT, Visakorpi T and
Maitland NJ: MicroRNA expression profile of primary prostate cancer
stem cells as a source of biomarkers and therapeutic targets. Eur
Urol. 67:7–10. 2015. View Article : Google Scholar
|
|
6
|
Xue Z, Wen J, Chu X and Xue X: A microRNA
gene signature for identification of lung cancer. Surg Oncol.
23:126–131. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yu J, Feng J, Zhi X, Tang J, Li Z, Xu Y,
Yang L, Hu Z and Xu Z: Let-7b inhibits cell proliferation,
migration, and invasion through targeting Cthrc1 in gastric cancer.
Tumour Biol. 36:3221–3229. 2015. View Article : Google Scholar
|
|
8
|
Volinia S, Nuovo G, Drusco A, Costinean S,
Abujarour R, Desponts C, Garofalo M, Baffa R, Aeqilan R, Maharry K,
et al: Pluripotent stem cell miRNAs and metastasis in invasive
breast cancer. J Natl Cancer Inst. 106:1062014. View Article : Google Scholar
|
|
9
|
Rudner J, Elsaesser SJ, Müller AC, Belka C
and Jendrossek V: Differential effects of anti-apoptotic Bcl-2
family members Mcl-1, Bcl-2, and Bcl-xL on celecoxib-induced
apoptosis. Biochem Pharmac. 79:10–20. 2010. View Article : Google Scholar
|
|
10
|
Aikawa T, Shinzawa K, Tanaka N and
Tsujimoto Y: Noxa is necessary for hydrogen peroxide-induced
caspase-dependent cell death. FEBS Lett. 584:681–688. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Cheng EHYA, Wei MC, Weiler S, Flavell RA,
Mak TW and Lindsten T: BCL-2, BCL-X(L) sequester BH3 domain-only
molecules preventing BAX- and BAK-mediated mitochondrial apoptosis.
Mol Cell. 8:705–711. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sobin LH, Gospodarowicz MK and Wittekind
C: TNM Classification of Malignant Tumors. 7th Edition.
Wiley-Blackwell; Oxford: 2010
|
|
13
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
14
|
Hong L, Han Y, Zhang Y, Zhang H, Zhao Q,
Wu K and Fan D: MicroRNA-21: A therapeutic target for reversing
drug resistance in cancer. Expert Opin Ther Targets. 17:1073–1080.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang Z, Han J, Cui Y, Fan K and Zhou X:
Circulating microRNA-21 as noninvasive predictive biomarker for
response in cancer immunotherapy. Med Hypotheses. 81:41–43. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Halimi M, Parsian H, Asghari SM, Sariri R,
Moslemi D, Yeganeh F and Zabihi E: Clinical translation of human
microRNA 21 as a potential biomarker for exposure to ionizing
radiation. Transl Res. 163:578–584. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Mace TA, Collins AL, Wojcik SE, Croce CM,
Lesinski GB and Bloomston M: Hypoxia induces the overexpression of
microRNA-21 in pancreatic cancer cells. J Surg Res. 184:855–860.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Tomimaru Y, Eguchi H, Nagano H, Wada H,
Kobayashi S, Marubashi S, Tanemura M, Tomokuni A, Takemasa I,
Umeshita K, et al: Circulating microRNA-21 as a novel biomarker for
hepatocellular carcinoma. Journal of Hepatology. 56:167–175. 2012.
View Article : Google Scholar
|
|
19
|
Wang ZX, Lu BB, Wang H, Cheng ZX and Yin
YM: MicroRNA-21 modulates chemosensitivity of breast cancer cells
to doxorubicin by targeting PTEN. Arch Med Res. 42:281–290. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hsu CC, Wu YC, Farh L, Du YC, Tseng WK, Wu
CC and Chang FR: Physalin B from Physalis angulata triggers the
NOXA-related apoptosis pathway of human melanoma A375 cells. Food
Chem Toxicol. 50:619–624. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Nakajima W and Tanaka N: Noxa induces
apoptosis in oncogene-expressing cells through catch-and-release
mechanism operating between Puma and Mcl-1. Biochem Biophys Res
Commun. 413:643–648. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Huskey NE, Guo T, Evason KJ, Momcilovic O,
Pardo D, Creasman KJ, Judson RL, Blelloch R, Oakes SA, Hebrok M and
Goga L: CDK1 Inhibition targets the p53-NOXA-MCL1 axis, selectively
kills embryonic stem cells, and prevents teratoma formation. Stem
Cell Reports. 4:374–389. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Leal AS, Wang R, Salvador JA and Jing Y:
Synthesis of novel ursolic acid heterocyclic derivatives with
improved abilities of antiproliferation and induction of p53,
p21waf1 and NOXA in pancreatic cancer cells. Bioorg Med Chem.
20:5774–5786. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Park SY, Jeong MS and Jang SB: In vitro
binding properties of tumor suppressor p53 with PUMA and NOXA.
Biochem Biophys Res Commun. 420:350–356. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Belmar J and Fesik SW: Small molecule
Mcl-1 inhibitors for the treatment of cancer. Pharmacol Ther.
145:76–84. 2015. View Article : Google Scholar :
|
|
26
|
Kuwahara Y, Wei D, Durand J and Weissman
BE: SNF5 reex-pression in malignant rhabdoid tumors regulates
transcription of target genes by recruitment of SWI/SNF complexes
and RNAPII to the transcription start site of their promoters. Mol
Cancer Res. 11:251–260. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ehrhardt H, Höfig I, Wachter F, Obexer P,
Fulda S, Terziyska N and Jeremias I: NOXA as critical mediator for
drug combinations in polychemotherapy. Cell Death Dis. 3:e3272012.
View Article : Google Scholar : PubMed/NCBI
|