Introduction
MicroRNAs, a novel class of endogenous small
non-coding RNAs, can modulate gene expression at the
post-translational level by interacting with the 3′ untranslated
regions of target mRNAs, resulting in the inhibited translation or
degradation of mRNAs (1,2). Substantial evidence has shown that
the majority of the regulated genes of microRNAs are important in
tumorigenesis, with aberrant microRNA expression reported in the
development and progression of several types of tumor (3,4). The
role of various microRNAs on invasion, migration and metastasis,
which are essential steps during cancer progression, has been
described (5–7).
Nasopharyngeal carcinoma (NPC) usually develops
around the ostium of the eustachian tube in the lateral wall of the
nasopharynx, and is widely prevalent in Southeast Asia, the Middle
East and North Africa (8). NPC is
also distinguished by its high rate of metastasis and poor
prognosis among head and neck cancer (9). Epstein-Barr virus (EBV) infection,
non-viral environmental risk factors and host genetics have been
generally accepted as three predominant factors, which contribute
to the development of NPC (10–12).
However, the molecular mechanism underlying the pathogenesis of NPC
remains to be fully elucidated.
Increasing evidence suggests that the distinct
expression pattern of microRNAs can provide important insight into
the molecular mechanism of tumorigenesis in NPC (13,14).
Several dysregulated miRNAs have been shown to regulate cell
growth, apoptosis and the metastasis of NPC (15–17).
Previous systematic investigations of microRNA expression profiles
in the stepwise development of NPC have also revealed 13 microRNAs,
which may be the most important modulators during the development
of NPC (18). MicroRNA-429
(miR-429), one of these microRNAs, has been reported to be
important in certain types of cancer. miR-429 can induce the
tumorigenesis of human non-small cell lung cancer and
mesenchymal-to-epithelial transition in metastatic ovarian cancer
cells (19,20). It can also inhibit cell invasion in
colorectal carcinoma, and the migration and invasion of breast
cancer cells, in which the expression profiles of miR-429 are
downregulated (21,22). However, the effects and possible
mechanisms of action of miR-429 in the metastasis of NPC have not
been examined.
In the present study, the expression levels of
miR-429 were detected in CNE-1 and CNE-2, which are two generally
used EBV-negative epithelial cells with different degrees of
differentiation (23,24). To improve understanding of the
regulatory mechanism of miR-429 in NPC, cell proliferation,
invasion and migration were analyzed in miR-429-overexpressing
CNE-2 cells. The modulatory function of miR-429 was also
investigated through two representative target genes, zinc finger
E-Box-binding homeobox 1 (ZEB1) and CRK-like (CRKL). The present
study aimed to investigate the potential function of miR-429 in NPC
tumorigenesis, using its target genes, including ZEB1 and CRKL, in
order to determine the potential application of miR-429 in NPC
treatment or prognosis determination.
Materials and methods
Cell lines and cell culture
NP69 cells, which are immortalized non-tumorigenic
nasopharyngeal epithelial cells, were cultured in
keratinocyte-serum-free medium (Invitrogen; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) supplemented with bovine
pituitary extract, as described previously (25). The two tumorigenic NPC cell lines,
comprising well-differentiated CNE-1 cells and
poorly-differentiated CNE-2 cells, were maintained in our
laboratory and cultured in RPMI-1640 (Invitrogen; Thermo Fisher
Scientific, Inc.) supplemented with 10% fetal bovine serum (BD
Biosciences; Franklin Lakes, NJ, USA). All cells were grown in a
humidified incubator at 37°C with 5% CO2. Following 24-h
incubation to 100% confluence, the cells were fixed with absolute
methanol (Sigma-Aldrich, St. Louis, MO, USA), stained with 0.4%
(w/v) crystal violet (Sigma-Aldrich) in methanol and subsequently
rinsed with water. Morphological images of three NPC cells were
captured using a Nikon Eclipse TS100 inverted light microscope
equipped with a Nikon Coolpix 4500 digital camera (Nikon
Corporation, Tokyo, Japan).
Transfection with microRNA mimics
The miR-429 mimic (with a nonspecific miRNA control)
and anti-miR429 (with a nonspecific anti-miRNA control) were all
purchased from Dharmacon; Thermo Fisher Scientific, Inc.). RNAiMAX
reagent (Invitrogen;Thermo Fisher Scientific, Inc.) was used to
deliver the miRNA mimics (20 nM) into the cells, which were
maintained in 6-well plates at a density of 1×106
cells/well, and transfection was performed using Lipofectamine 2000
(Invitrogen;Thermo Fisher Scientific, Inc.), according to the
manufacturer's protocol. Transfected cells were incubated at 37°C
in complete medium, and the subsequent experiments were performed
48 h following transfection.
RNA extraction and reverse
transcription
Total RNA was extracted from the cells using TRIzol
(Invitrogen;Thermo Fisher Scientific, Inc.), according to the
manufacturer's protocol. Briefly, cells at ~80% confluence were
seeded into 6-well plates and washed twice using ice-cold
phosphate-buffered saline (PBS), following which 1 ml TRIzol was
added to obtain the RNA. The quantity and quality of the extracted
RNA were analyzed using a Nanodrop ND1000 spectrophotometer (Thermo
Fisher Scientific, Inc.). For mRNA analysis, cDNA was synthesized
using Quant cDNA with random primers (Tiangen Biotech Co., Ltd.,
Beijing, China), and the miRNAs were reverse-transcribed, according
to previously described method (26). Briefly, a specific reverse primer
targeting individual miRNA was designed to complete the reverse
transcription, and one miRNA-specific forward primer and one
universal reverse primer were used in the subsequent quantitative
polymerase chain reaction (qPCR) analysis.
qPCR
The qPCR detection was completed using SYBR Premix
Ex Taq™ II (Takara Bio, Inc., Otsu, Japan), according to the
manufacturer's protocol. The qPCR procedure was performed in
accordance with the protocol of Takara Bio, Inc. in a StepOne Plus
Real-time PCR system (Applied Biosystems; Thermo Fisher Scientific,
Inc.). The results were analyzed using the 2−∆∆Cq method
(27). U6 small nuclear RNA
(U6-snRNA; Guangzhou RiboBio Co., Ltd., Guangzhou, China) and
β-actin were used as internal controls for microRNA and mRNA,
respectively. The primers used are listed in Table I.
 | Table IPrimers used in quantitative
polymerase chain reaction analysis. |
Table I
Primers used in quantitative
polymerase chain reaction analysis.
| Primer | Sequence
(5′-3′) |
|---|
| miR-429-R |
CTCAACTGGTGTCGTGGAGTCGG
CAATTCAGTTGAGACGGTTTT |
| miR-429-F |
ACACTCCAGCTGGGTAATACTGTC TGGTAA |
| Universal-R |
TGGTGTCGTGGAGTCG |
| U6-F |
CTCGCTTCGGCAGCACA |
| U6-R |
AACGCTTCACGAATTTGCGT |
| ZEB1-F |
GCACAACCAAGTGCAGAAGA |
| ZEB1-R |
GCCTGGTTCAGGAGAAGATG |
| CRKL-F |
CGCTCCGCCTGGTATATGG |
| CRKL-R |
GGACACCGACAGCACATAGTC |
| β-actin-F |
AGTGTGACGTGGACATCCGCA |
| β-actin-R |
ATCCACATCTGCTGGAAGGTGGAC |
Cell proliferation assays
Freshly sorted NP69, CNE-1 and CNE-2 cells were
incubated at a density of 500 cells per well in a 96-well plate in
triplicate to examine the growth rate. The cells transfected with
miRNAs were reseeded at a density of 1.5×103 cells/well
in a final volume of 150 µl 48 h following incubation, and
incubated at 37°C overnight. The effects of miR-429 on cell growth
and proliferation were determined using an MTT assay
(Sigma-Aldrich), as described previously (28). During the subsequent 4 days, the
absorbance of the cells stained with 50 µl MTT were measured
at 570 nm using a Multiskan MK3 microplate reader (Thermo Fisher
Scientific, Inc.), and the measurements were used to construct a
cell growth curve.
Cell invasion assays
Cell invasion was measured using Biocoat Matrigel
Invasion Chambers (BD Biosciences), according to the manufacturer's
protocol. In brief, the CNE-2 cells transfected with the miRNA
mimics or inhibitors were plated 48 h post-transfection in
serum-free medium (2.5×104 cells per Transwell) and
allowed to migrate towards a 10% fetal bovine serum gradient for 12
h at 37°C. Subsequently, the upper chambers were removed from the
lower chambers and wiped using cotton swabs. The invaded cells were
fixed using ≥99.9% methanol (Sigma-Aldrich), and visualized by 0.1%
toluidine blue staining (Sigma-Aldrich) under a Nikon Eclipse TS100
light microscope, as described in a previous study (16). This experiment was independently
repeated at least twice.
Western blot analysis
Western blot analysis was performed, as described
previously (29). In brief, the
cells were carefully collected with scrapers on ice and then
subjected to lysis with radioimmunoprecipitation buffer (Beyotime
Institute of Biotechnology, Nanjing, China). Protein lysates were
separated by 12% SDS-PAGE (Beyotime Institute of Biotechnology) and
subsequently electrophoretically transferred onto a polyvinylidene
difluoride membrane (EMD Millipore, Billerica, MA, ISA).
Tris-buffered saline with Tween-20 (TBS-T) buffer, containing 10 mM
Tris-HCl (pH 7.5; Beyotime), 150 mM NaCl (GR grade; Shanghai Hushi,
China) and 0.1 % (v/v) Tween-20 (Sigma-Aldrich), was pre-prepared
for use in the washing and blocking steps and was used as the
antibody diluent. Following blocking with 5% (v/v) skimmed milk in
TBS-T buffer for 1 h at room temperature, protein expression levels
were analyzed following incubation with the following primary
antibodies overnight at 4°C: Polyclonal rabbit anti-ZEB1 (1:1,000;
HPA027524; Sigma-Aldrich), monoclonal rabbit anti-CRKL (1:500;
Y244; Abcam, Cambridge, UK) and monoclonal mouse anti-β-actin
(1:5,000; A5441; Sigma-Aldrich). After rinsing three times with
TBS-T, the membranes were incubated with secondary horseradish
peroxidase-conjugated anti-rabbit antibody (7074) or anti-mouse
antibodies (both 1:2,000; 7076, both Cell Signaling Technology,
Inc., Danvers, MA, USA) at room temperature for 60 min. Following
washing in TBS-T, images of the immunoblots were acquired and
analyzed using an ImageQuant LAS 4000 system (GE Healthcare
Bio-Sciences, Pittsburgh, PA, USA). The primary antibodies used in
the experiment were ZEB1, CRKL and β-actin.
Statistical analysis
The results of the quantitative data in the present
study are expressed as the mean ± standard deviation. Two-tailed
Student's t-test was used for comparisons of two independent
groups, and Welch's corrected t-test was used for unequal
variances. SPSS 13.0 software (SPSS, Inc., Chicago, IL, USA) was
used for statistical analysis. P<0.05 was considered to indicate
a statistically significant difference.
Results
Expression of miR-429 is decreased in
human NPC cell lines
As an initial step in assessing the potential role
of miR-429 in the development of NPC, the expression profiles were
analyzed in two NPC-derived cell lines, CNE-l and CNE-2, with
different levels of differentiation. The immortalized NPC cells
(NP69) were used as a negative control. The three carcinoma cell
lines were incubated and observed under a Nikon Eclipse TS100 light
microscope to confirm their morphologies (Fig. 1A). Further measurement of cell
proliferation showed that CNE-1 and CNE-2 exhibited higher growth
rates, compared with the NP69 cells. In addition, the CNE-2 cells
exhibited higher proliferation rates, compared with the CNE-1
(Fig. 1B), indicating higher
malignancy potential. Of note, the expression of miR-429 was
suppressed more significantly in the CNE-2 cells, compared with the
CNE-1 cells (Fig. 1C). CNE-2 is
derived from poorly-differentiated NPC cells (24), and exhibits higher epidemicity and
malignancy, compared with well-differentiated NPC cells, including
CNE-1 cells (23). The results of
the present study showed aberrant miR-429 expression in the
low-differentiated NPC cells, indicating that miR-429 may be more
important in the pathogenesis of NPC. The CNE-2 cells were selected
to further investigate the functional role of miR-429 in NPC
tumorigenesis.
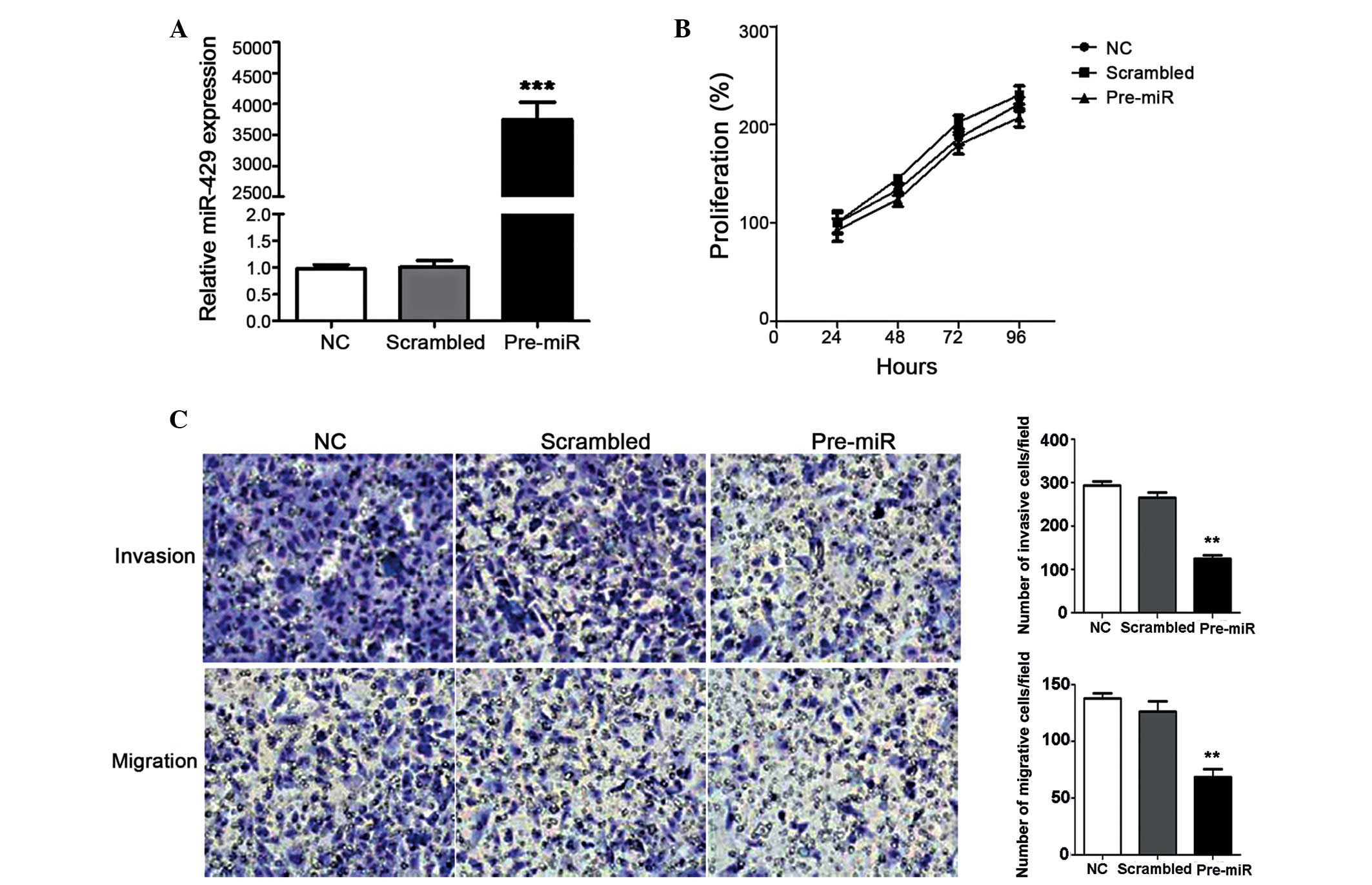
Overexpression of miR-429 suppresses cell
invasion and migration
In order to explore the functional roles of miR-429,
miR-429 overexpression was induced in CNE-2 cells by transfection
with miR-429 mimics, and the effects on cell proliferation,
migration and invasion were investigated. Overexpression of miR-429
in CNE-2 cells was confirmed using RT-qPCR 48 h after transfection
to ensure the induction had occurred (Fig. 2A), which demonstrated a significant
3,700-fold increase. The results indicated that the overexpression
of miR-429 had minimal effect on cell proliferation over a 96-h
period of detection (Fig. 2B).
However, the miR-429-overexpressing cells demonstrated decreased
invasion and migration, compared with the control group and the
scrambled cells, which were transfected with nonspecific miRNA
(Fig. 2C). Cell invasion was
suppressed more markedly. Taken together, miR-429 may have
regulated NPC tumorigenesis in a negative manner, indicating its
potential in miRNA-based therapy against NPC. For further
explanation of this negative regulation, target genes of miR-429
were also investigated.
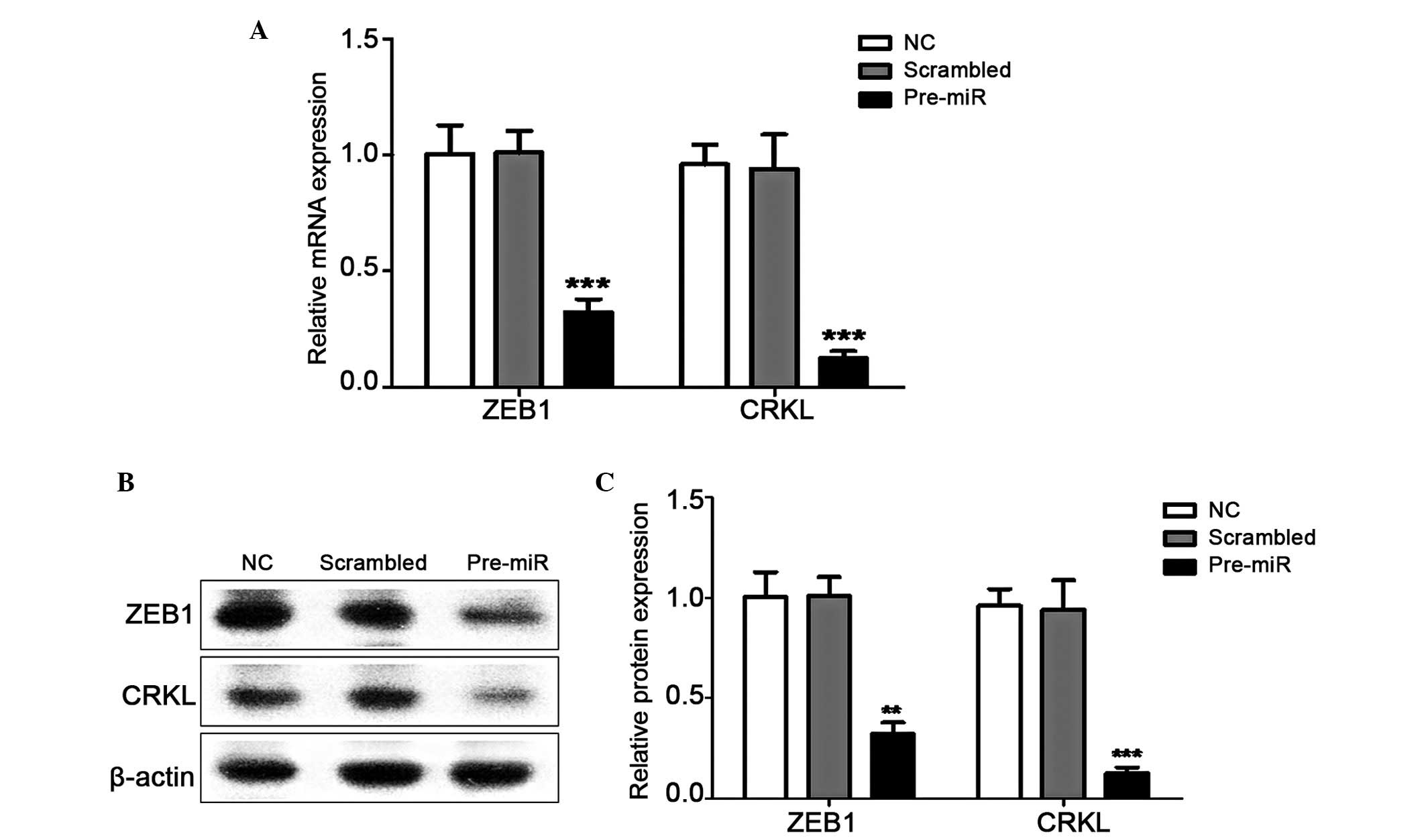
miR-429 inhibits the expression levels of
ZEB1 and CRKL
ZEB1, which is an important
epithelial-to-mesenchymal transition (EMT) inducer (30), and CRKL, which has been identified
as a candidate target of miR-429 (22), were selected in the present study
as representatives to examine the regulator function of miR-429 in
NPC cells.
The mRNA and protein expression levels of ZEB1 and
CRKL were detected in response to the induced expression of miR-429
in CNE-2 cells by transfection with miR429 mimics. The results
showed that ZEB1 and CRKL were suppressed significantly in the
miR-429-overexpressing cells (Fig.
3). Compared with the scrambled cells, the expression levels of
ZEB1 and CRKL were downregulated by ~3-fold and 6-fold,
respectively in the pre-miR transfected cells at the mRNA and
protein levels. These results indicated that miR-429 may regulate
the development of NPC by inhibiting the function of its target
genes.
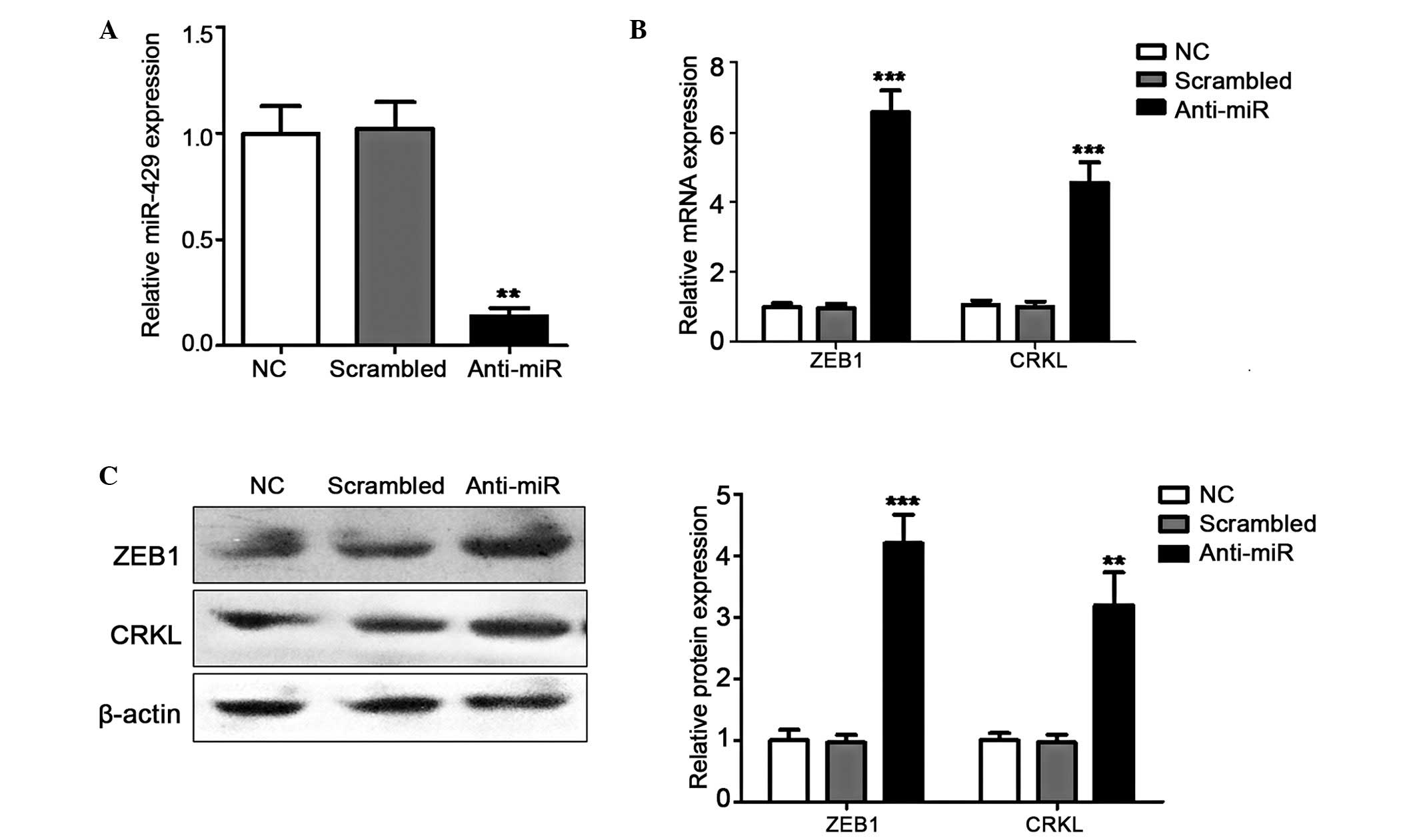
miR-429 silencing induces the expression
levels of ZEB1 and CRKL
The gain-of-function investigated described above
indicated that ZEB1 and CRKL were suppressed by miR429. To verify
this negative regulation, their relative expression levels were
also investigated in miR-429-silenced cells through anti-miR429
transfection. miR-429 silencing was confirmed using RT-qPCR
(Fig. 4A), and the expression
levels of ZEB1 and CRKL were detected as described above. As
expected, the expression levels of the two target genes were
restored and even induced by the downregulation of miR-429 at the
mRNA and protein levels (Fig. 4B and
C). These results concluded that ZEB1 and CRKL were negatively
regulated by miR429 in the NPC cells. Notably, miR-429 may suppress
cell motility in NPC by negatively modulating its target genes,
including ZEB1 and CRKL, indicating its potential as a candidate
for miRNA-based prognosis or therapy against NPC.
Discussion
NPC has distinct ethnic and geographic
distributions, and is particularly common in the southern Chinese
population (31). The mechanism of
NPC tumorigenesis is complex, involving aberrations in a variety of
pathways and alterations in the expression levels of several
proteins (32). Although high
survival rates are reported for early stage NPC, the majority of
NPC cases are diagnosed at an advanced stage. The prognosis for
metastatic disease remains poor due to delays in seeking treatment
following the onset of symptoms, although a thorough nasopharyngeal
examination is difficult to complete (33). Therefore, identifying effective
diagnostic biomarkers and targeted treatments for NPC is essential
to improve the clinical outcomes. Differentially expressed miRNAs
have been screened out for candidate biomarkers in NPC (34–36).
miR-429 is a member of the miR-200 family, and four
members of this family have been found to be important in the
regulation of EMT in various types of tumor (37). In the present study, miR-429 was
markedly downregulated in poorly-differentiated CNE-2 cells
(Fig. 1), in accordance with a
previous microRNA microarray study (18). Further investigations indicated
that the overexpression of miR-429 inhibited the cell migration and
invasion of NPC cells in vitro (Fig. 2), which was also in accordance with
previously reported results in other types of carcinoma (21,22,38).
The upregulation of miR-429 inhibits invasion and promotes
apoptosis in esophageal carcinoma cells by targeting B cell
lymphoma-2 and SP1 (38). As
described in breast cancer cells, miR-429 can suppress cell
motility by negatively modulating several key invasion and
metastasis inducers, including ZEB1 and CRKL. ZEB1, also known as
δEF1, can repress the transcription of E-cadherin and regulate
epithelial plasticity in breast cancer cells (39). CRKL, a tyrosine-phosphorylated
protein, can transform fibroblasts and function in transformation
via the BCR-ABL oncogene (40). In
addition, CRKL is important in proliferation, migration and the
evasion of apoptosis (22).
Downregulation in the levels of ZEB1 and CRKL were detected in the
miR-429-overexpressing CNE-2 cells (Fig. 3). In addition, the downregulation
of miR-429 led to a reversal in the promoted expression profiles of
ZEB1 and CRKL (Fig. 4). These
results indicated that repression in the invasion and migration of
miR-429-overexpressing NPC cells was closely associated with the
functions of target genes, including ZEB1 and CRKL, regulated by
miR-429.
In conclusion, significant changes in the expression
of miR-429 were detected, particularly in low-differentiated CNE-2
cells. Further results showed that miR-429 inhibited the invasion
and migration of CNE-2 cells. In addition, the mRNA and protein
expression levels of the two target genes, ZEB1 and CRKL, were
downregulated and upregulated by transfection with the miR429 mimic
and anti-miR429, respectively. These results indicated that miR-429
may suppress cell motility in NPC by negatively modulating its
target genes, including ZEB1 and CRKL. Therefore, miR-429 has
potential for use as a biomarker of EMT in NPC, and also has
potential therapeutic value in abating NPC metastasis, particularly
in undifferentiated NPC cells. Further investigations may assist in
clarifying the complex mechanisms of miR-429 regulation in NPC
metastasis for improving prognosis and therapy.
References
|
1
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Schirle NT, Sheu-Gruttadauria J and MacRae
IJ: Structural basis for microRNA targeting. Science. 346:608–613.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lu J, Getz G, Miska EA, Alvarez Saavedra
E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA,
et al: MicroRNA expression profiles classify human cancers. Nature.
435:834–838. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Xie YJ, Long ZF and He XS: Involvement of
EBV-encoded BART-miRNAs and dysregulated cellular miRNAs in
naso-pharyngeal carcinoma genesis. Asian Pac J Cancer Prev.
14:5637–5644. 2013. View Article : Google Scholar
|
|
5
|
Buffa FM, Camps C, Winchester L, Snell CE,
Gee HE, Sheldon H, Taylor M, Harris AL and Ragoussis J:
microRNA-associated progression pathways and potential therapeutic
targets identified by integrated mRNA and microRNA expression
profiling in breast cancer. Cancer Res. 71:5635–5645. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bojmar L, Karlsson E, Ellegård S, Olsson
H, Björnsson B, Hallböök O, Larsson M, Stål O and Sandström P: The
role of microRNA-200 in progression of human colorectal and breast
cancer. PloS One. 8:e848152013. View Article : Google Scholar :
|
|
7
|
Baranwal S and Alahari SK: miRNA control
of tumor cell invasion and metastasis. Int J Cancer. 126:1283–1290.
2010.
|
|
8
|
Brennan B: Nasopharyngeal carcinoma.
Orphanet J Rare Dis. 1:232006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lo KW, To KF and Huang DP: Focus on
nasopharyngeal carcinoma. Cancer Cell. 5:423–428. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lung ML: Unlocking the rosetta stone
enigma for naso-pharyngeal carcinoma: Genetics, viral infection and
epidemiological factors. Semin Cancer Biol. 22:77–78. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Nor Hashim NA, Ramzi NH, Velapasamy S,
Alex L, Chahil JK, Lye SH, Munretnam K, Haron MR and Ler LW:
Identification of genetic and non-genetic risk factors for
naso-pharyngeal carcinoma in a Southeast Asian population. Asian
Pac J Cancer Prev. 13:6005–6010. 2012. View Article : Google Scholar
|
|
12
|
Hildesheim A and Wang CP: Genetic
predisposition factors and nasopharyngeal carcinoma risk: A review
of epidemiological association studies, 2000–2011: Rosetta Stone
for NPC: Genetics, viral infection and other environmental factors.
Semin Cancer Biol. 22:107–116. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Liu N, Chen NY, Cui RX, Li WF, Li Y, Wei
RR, Zhang MY, Sun Y, Huang BJ, Chen M, et al: Prognostic value of a
microRNA signature in nasopharyngeal carcinoma: A microRNA
expression analysis. Lancet Oncol. 13:633–641. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yu BL, Peng XH, Zhao FP, Liu X, Lu J, Wang
L, Li G, Chen HH and Li XP: MicroRNA-378 functions as an onco-miR
in nasopharyngeal carcinoma by repressing TOB2 expression. Int J
Oncol. 44:1215–1222. 2014.PubMed/NCBI
|
|
15
|
Deng M, Tang H, Zhou Y, Zhou M, Xiong W,
Zheng Y, Ye Q, Zeng X, Liao Q, Guo X, et al: miR-216b suppresses
tumor growth and invasion by targeting KRAS in nasopharyngeal
carcinoma. J Cell Sci. 124:2997–3005. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Xia H, Ng SS, Jiang S, Cheung WK, Sze J,
Bian XW, Kung HF and Lin MC: miR-200a-mediated downregulation of
ZEB2 and CTNNB1 differentially inhibits nasopharyngeal carcinoma
cell growth, migration and invasion. Biochem Biophys Res Commun.
391:535–541. 2010. View Article : Google Scholar
|
|
17
|
Sun XJ, Liu H, Zhang P, Zhang XD, Jiang ZW
and Jiang CC: miR-10b promotes migration and invasion in
nasopharyngeal carcinoma cells. Asian Pac J Cancer Prev.
14:5533–5537. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Luo Z, Zhang L, Li Z, Li X and Li G, Yu H,
Jiang C, Dai Y, Guo X, Xiang J and Li G: An in silico analysis of
dynamic changes in microRNA expression profiles in stepwise
development of naso-pharyngeal carcinoma. BMC Med Genomics.
5:32012. View Article : Google Scholar
|
|
19
|
Chen J, Wang L, Matyunina LV, Hill CG and
McDonald JF: Overexpression of miR-429 induces
mesenchymal-to-epithelial transition (MET) in metastatic ovarian
cancer cells. Gynecol Oncol. 121:200–205. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhu W, He J, Chen D, Zhang B, Xu L, Ma H,
Liu X, Zhang Y and Le H: Expression of miR-29c, miR-93 and miR-429
as potential biomarkers for detection of early stage non-small lung
cancer. PloS One. 9:e877802014. View Article : Google Scholar
|
|
21
|
Sun Y, Shen S, Liu X, Tang H, Wang Z, Yu
Z, Li X and Wu M: MiR-429 inhibits cells growth and invasion and
regulates EMT-related marker genes by targeting Onecut2 in
colorectal carcinoma. Mol Cell Biochem. 390:19–30. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ye ZB, Ma G, Zhao YH, Xiao Y, Zhan Y, Jing
C, Gao K, Liu ZH and Yu SJ: miR-429 inhibits migration and invasion
of breast cancer cells in vitro. Int J Oncol. 46:531–538. 2015.
|
|
23
|
Zhang S, Wu Y, Zeng Y, Zech L and Klein G:
Cytogenetic studies on an epithelioid cell line derived from
nasopharyngeal carcinoma. Hereditas. 97:23–28. 1982. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sizhong Z, Xiukung G and Yi Z: Cytogenetic
studies on an epithelial cell line derived from poorly
differentiated nasopharyngeal carcinoma. Int J Cancer. 31:587–590.
1983. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Tsao SW, Wang X, Liu Y, Cheung YC, Feng H,
Zheng Z, Wong N, Yuen PW, Lo AK, Wong YC and Huang DP:
Establishment of two immortalized nasopharyngeal epithelial cell
lines using SV40 large T and HPV16E6/E7 viral oncogenes. Biochim
Biophys Acta. 1590:150–158. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee
DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, et al:
Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic
Acids Res. 33:e1792005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Schmittgen TD and Livak KJ: Analyzing
real-time PCR data by the comparative C(T) method. Nat Protoc.
3:1101–1108. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Tsang WP and Kwok TT: The
miR-18a* microRNA functions as a potential tumor
suppressor by targeting on K-Ras. Carcinogenesis. 30:953–959. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Liu X, Lv XB, Wang XP, Sang Y, Xu S, Hu K,
Wu M, Liang Y, Liu P, Tang J, et al: MiR-138 suppressed
nasopharyngeal carcinoma growth and tumorigenesis by targeting the
CCND1 oncogene. Cell Cycle. 11:2495–2506. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Peinado H, Olmeda D and Cano A: Snail, Zeb
and bHLH factors in tumour progression: An alliance against the
epithelial phenotype? Nat Rev Cancer. 7:415–428. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yu MC and Yuan JM: Epidemiology of
nasopharyngeal carcinoma. Semin Cancer Biol. 12:421–429. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Chou J, Lin YC, Kim J, You L, Xu Z, He B
and Jablons DM: Nasopharyngeal carcinoma-review of the molecular
mechanisms of tumorigenesis. Head Neck. 30:946–963. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lee AW, Foo W, Law SC, Poon YF, Sze WM, O
SK, Tung SY and Lau WH: Nasopharyngeal carcinoma: presenting
symptoms and duration before diagnosis. Hong Kong Med J. 3:355–361.
1997.
|
|
34
|
Zeng Z, Zhou Y, Xiong W, Luo X, Zhang W,
Li X, Fan S, Cao L, Tang K, Wu M and Li G: Analysis of gene
expression identifies candidate molecular markers in nasopharyngeal
carcinoma using microdissection and cDNA microarray. J Cancer Res
Clin Oncol. 133:71–81. 2007. View Article : Google Scholar
|
|
35
|
Li G, Liu Y, Su Z, Ren S, Zhu G, Tian Y
and Qiu Y: MicroRNA-324–3p regulates nasopharyngeal carcinoma
radioresistance by directly targeting WNT2B. Eur J Cancer.
49:2596–2607. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Lu J, Xu X, Liu X, Peng Y, Zhang B, Wang
L, Luo H, Peng X, Li G, Tian W, et al: Predictive value of miR-9 as
a potential biomarker for nasopharyngeal carcinoma metastasis. Br J
Cancer. 110:392–398. 2014. View Article : Google Scholar :
|
|
37
|
Korpal M and Kang Y: The emerging role of
miR-200 family of microRNAs in epithelial-mesenchymal transition
and cancer metastasis. RNA Biol. 5:115–119. 2008. View Article : Google Scholar
|
|
38
|
Wang Y, Li M, Zang W, Ma Y, Wang N, Li P,
Wang T and Zhao G: MiR-429 up-regulation induces apoptosis and
suppresses invasion by targeting Bcl-2 and SP-1 in esophageal
carcinoma. Cell Oncol (Dordr). 36:385–394. 2013. View Article : Google Scholar
|
|
39
|
Eger A, Aigner K, Sonderegger S, Dampier
B, Oehler S, Schreiber M, Berx G, Cano A, Beug H and Foisner R:
DeltaEF1 is a transcriptional repressor of E-cadherin and regulates
epithelial plasticity in breast cancer cells. Oncogene.
24:2375–2385. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Senechal K, Halpern J and Sawyers CL: The
CRKL adaptor protein transforms fibroblasts and functions in
transformation by the BCR-ABL oncogene. J Biol Chem.
271:23255–23261. 1996. View Article : Google Scholar : PubMed/NCBI
|