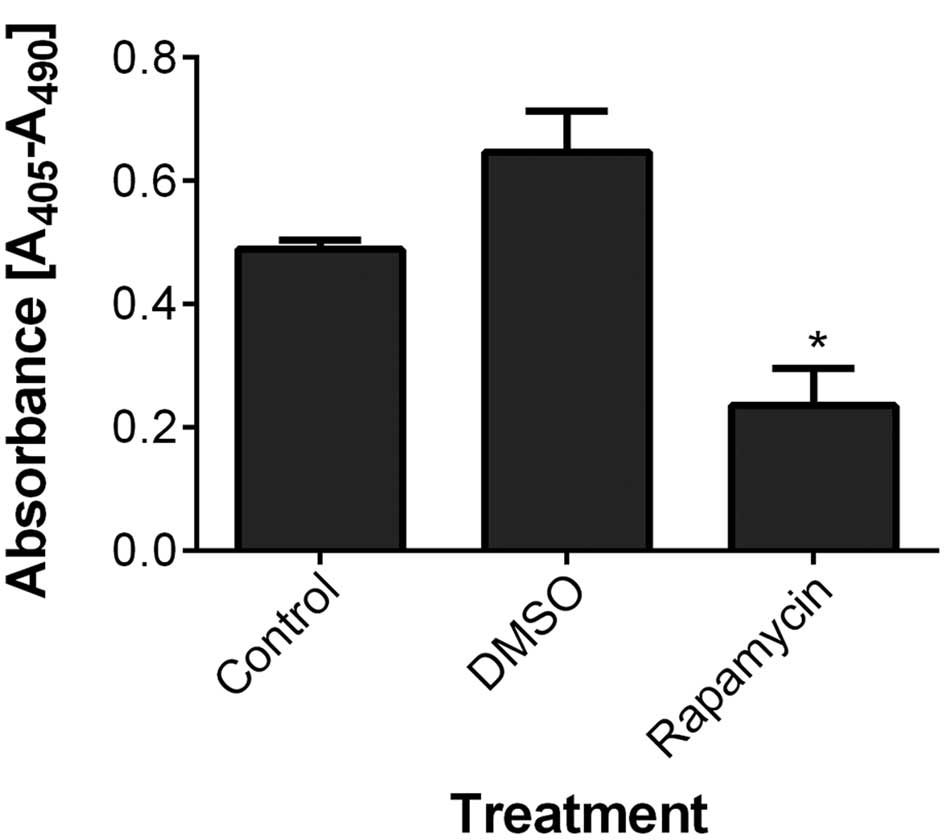
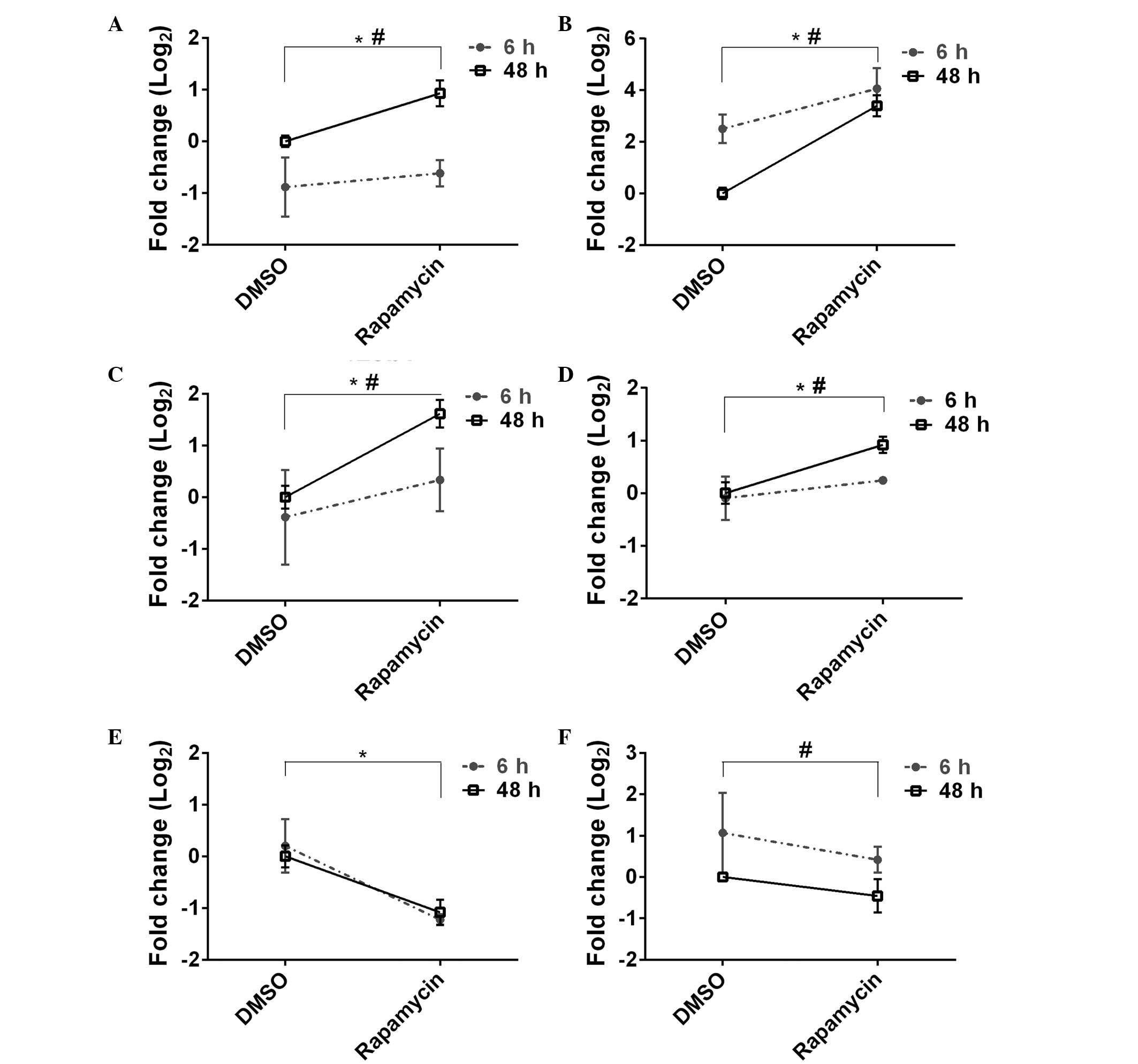
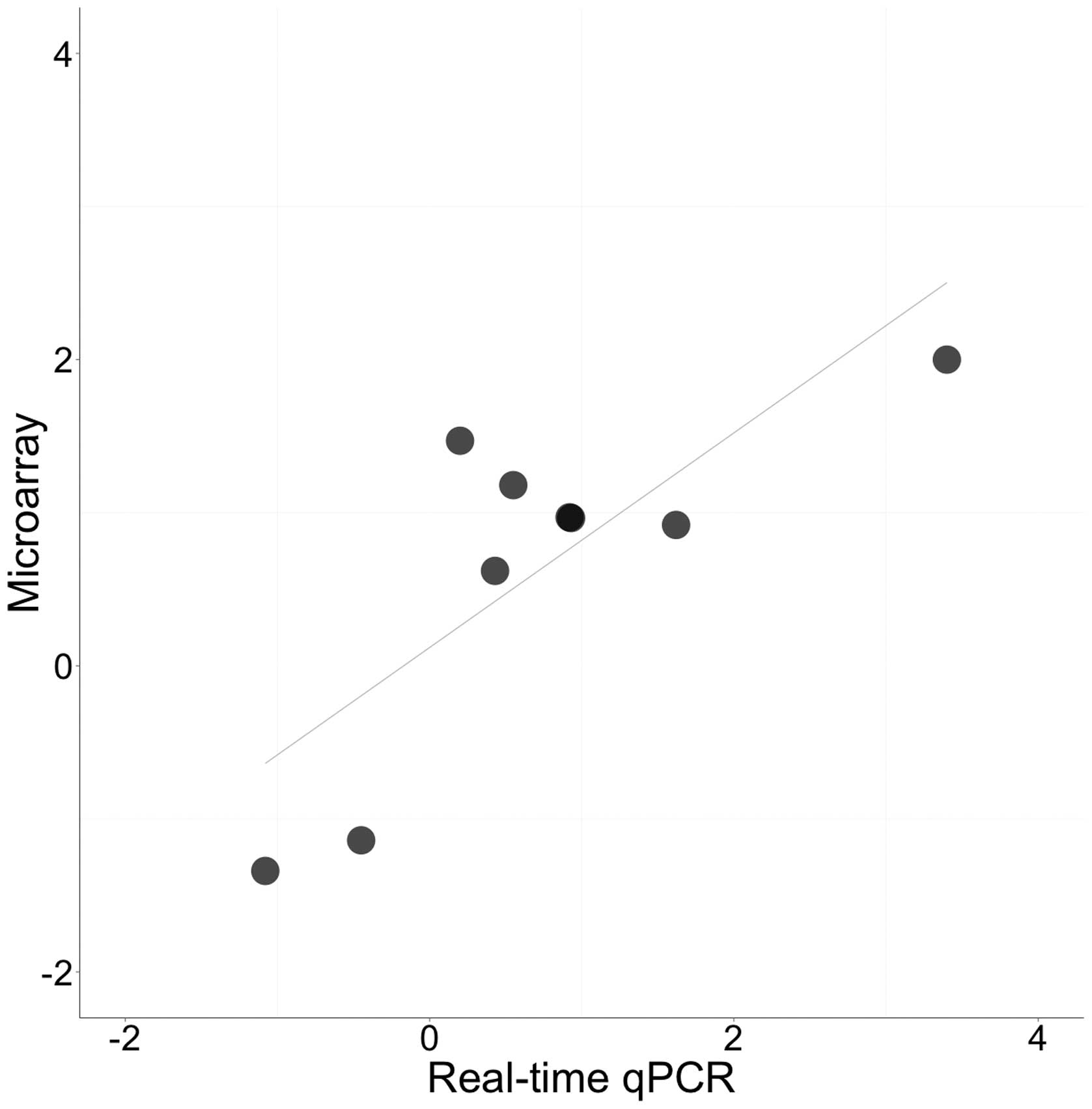
|
1
|
Corradetti MN and Guan KL: Upstream of the
mammalian target of rapamycin: Do all roads pass through mTOR?
Oncogene. 25:6347–6360. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Brown EJ, Albers MW, Shin TB, Ichikawa K,
Keith CT, Lane WS and Schreiber SL: A mammalian protein targeted by
G1-arresting rapamycin-receptor complex. Nature. 369:756–758. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Akcakanat A, Singh G, Hung MC and
Meric-Bernstam F: Rapamycin regulates the phosphorylation of
rictor. Biochem Biophys Res Commun. 362:330–333. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Rao RD, Buckner JC and Sarkaria JN:
Mammalian target of rapamycin (mTOR) inhibitors as anti-cancer
agents. Curr Cancer Drug Targets. 4:621–635. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Loewith R, Jacinto E, Wullschleger S,
Lorberg A, Crespo JL, Bonenfant D, Oppliger W, Jenoe P and Hall MN:
Two TOR complexes, only one of which is rapamycin sensitive, have
distinct roles in cell growth control. Mol Cell. 10:457–468. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Makky K, Tekiela J and Mayer AN: Target of
rapamycin (TOR) signaling controls epithelial morphogenesis in the
vertebrate intestine. Dev Biol. 303:501–513. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ji J and Zheng PS: Activation of mTOR
signaling pathway contributes to survival of cervical cancer cells.
Gynecol Oncol. 117:103–108. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Jimenez RH, Lee JS, Francesconi M,
Castellani G, Neretti N, Sanders JA, Sedivy J and Gruppuso PA:
Regulation of gene expression in hepatic cells by the mammalian
Target of Rapamycin (mTOR). PLoS One. 5:e90842010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Preiss T, Baron-Benhamou J, Ansorge W and
Hentze MW: Homodirectional changes in transcriptome composition and
mRNA translation induced by rapamycin and heat shock. Nat Struct
Biol. 10:1039–1047. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Cunningham JT, Rodgers JT, Arlow DH,
Vazquez F, Mootha VK and Puigserver P: mTOR controls mitochondrial
oxidative function through a YY1-PGC-1alpha transcriptional
complex. Nature. 450:736–740. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Duvel K, Yecies JL, Menon S, Raman P,
Lipovsky AI, Souza AL, Triantafellow E, Ma Q, Gorski R, Cleaver S,
et al: Activation of a metabolic gene regulatory network downstream
of mTOR complex 1. Mol Cell. 39:171–183. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Totary-Jain H, Sanoudou D, Ben-Dov IZ,
Dautriche CN, Guarnieri P, Marx SO, Tuschl T and Marks AR:
Reprogramming of the microRNA transcriptome mediates resistance to
rapamycin. J Biol Chem. 288:6034–6044. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hentges KE, Sirry B, Gingeras AC,
Sarbassov D, Sonenberg N, Sabatini D and Peterson AS: FRAP/mTOR is
required for proliferation and patterning during embryonic
development in the mouse. Proc Natl Acad Sci USA. 98:13796–13801.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Oldham S, Montagne J, Radimerski T, Thomas
G and Hafen E: Genetic and biochemical characterization of dTOR,
the Drosophila homolog of the target of rapamycin. Genes Dev.
14:2689–2694. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ganesan S, Moussavi Nik SH, Newman M and
Lardelli M: Identification and expression analysis of the zebrafish
orthologues of the mammalian MAP1LC3 gene family. Exp Cell Res.
328:228–237. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sapp V, Gaffney L, EauClaire SF and
Matthews RP: Fructose leads to hepatic steatosis in zebrafsih that
is reversed by mechanistic target of rapamycin (mTOR) inhibition.
Hepatology. 60:1581–1592. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Burkhalter MD, Fralish GB, Premont RT,
Caron MG and Philipp M: Grk5l controls heart development by
limiting mTOR signaling during symmetry breaking. Cell Rep.
4:625–632. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kim SH, Scott SA, Bennett MJ, Carson RP,
Fessel J, Brown HA and Ess KC: Multi-organ abnormalities and mTORC1
activation in zebrafish model of multiple acyl-CoA dehydrogenase
deficiency. PLoS Genet. 9:e10035632013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Xu B, Lee KK, Zhang L and Gerton JL:
Stimulation of mTORC1 with L-leucine rescues defects associated
with Roberts syndrome. PLoS Genet. 9:e10038572013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li Y, Huang W, Huang S, Du J and Huang C:
Screening of anti-cancer agent using zebrafish: Comparison with the
MTT assay. Biochem Biophys Res Commun. 422:85–90. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Driever W and Rangini Z: Characterization
of a cell line derived from zebrafish (Brachydanio rerio) embryos.
In Vitro Cell Dev Biol Anim. 29A:749–754. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Krishan A: Rapid flow cytofluorometric
analysis of mammalian cell cycle by propidium iodide staining. J
Cell Biol. 66:188–193. 1975. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Gautier L, Cope L, Bolstad BM and Irizarry
RA: Affy-analysis of Affymetrix GeneChip data at the probe level.
Bioinformatics. 20:307–315. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Bolstad BM, Collin F, Brettschneider J,
Simpson K, Cope L, Irizarry RA and Speed TP: Quality assessment of
affymetrix GeneChip data. Bioinformatics and Computational Biology
Solutions Using R and Bioconductor. Gentleman R, Carey V, Huber W,
Irizarry R and Dudoit S: Springer; New York: pp. 33–47. 2005,
View Article : Google Scholar
|
|
25
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Huang da W, Sherman BT and Lempicki RA:
Bioinformatics enrichment tools: Paths toward the comprehensive
functional analysis of large gene lists. Nucleic Acids Res.
37:1–13. 2009. View Article : Google Scholar
|
|
27
|
Wang X, Kang DD, Shen K, Song C, Lu S,
Chang LC, Liao SG, Huo Z, Tang S, Ding Y, et al: An R package suite
for microarray meta-analysis in quality control, differentially
expressed gene analysis and pathway enrichment detection.
Bioinformatics. 28:2534–2536. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
McCurley AT and Callard GV:
Characterization of housekeeping genes in zebrafish: Male-female
differences and effects of tissue type, developmental stage and
chemical treatment. BMC Mol Biol. 9:1022008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Pfaffl MW: A new mathematical model for
relative quantification in real-time RT-PCR. Nucleic Acids Res.
29:e452001. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Fay MP: Confidence intervals that match
Fisher's exact or Blaker's exact tests. Biostatistics. 11:373–374.
2010. View Article : Google Scholar :
|
|
31
|
Yildiz G, Arslan-Ergul A, Bagislar S, Konu
O, Yuzugullu H, Gursoy-Yuzugullu O, Ozturk N, Ozen C, Ozdag H,
Erdal E, et al: Genome-wide transcriptional reorganization
associated with senescence-to-immortality switch during human
hepatocellular carcinogenesis. PLoS One. 8:e640162013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wright GW and Simon RM: A random variance
model for detection of differential gene expression in small
microarray experiments. Bioinformatics. 19:2448–2455. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Terada N, Patel HR, Takase K, Kohno K,
Nairn AC and Gelfand EW: Rapamycin selectively inhibits translation
of mRNAs encoding elongation factors and ribosomal proteins. Proc
Natl Acad Sci USA. 91:11477–11481. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Grolleau A, Bowman J, Pradet-Balade B,
Puravs E, Hanash S, Garcia-Sanz JA and Beretta L: Global and
specific translational control by rapamycin in T cells uncovered by
microarrays and proteomics. J Biol Chem. 277:22175–22184. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Parent R, Kolippakkam D, Booth G and
Beretta L: Mammalian target of rapamycin activation impairs
hepatocytic differentiation and targets genes moderating lipid
homeostasis and hepatocellular growth. Cancer Res. 67:4337–4345.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Moore MJ: From birth to death: The complex
lives of eukaryotic mRNAs. Science. 309:1514–1518. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Boshoff HI, Myers TG, Copp BR, McNeil MR,
Wilson MA and Barry CE III: The transcriptional responses of
Mycobacterium tuberculosis to inhibitors of metabolism: Novel
insights into drug mechanisms of action. J Biol Chem.
279:40174–40184. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Bhat KP, Itahana K, Jin A and Zhang Y:
Essential role of ribosomal protein L11 in mediating growth
inhibition-induced p53 activation. EMBO J. 23:2402–2412. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Ravikumar B, Berger Z, Vacher C, O'Kane CJ
and Rubinsztein DC: Rapamycin pre-treatment protects against
apoptosis. Hum Mol Genet. 15:1209–1216. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Tirado OM, Mateo-Lozano S and Notario V:
Rapamycin induces apoptosis of JN-DSRCT-1 cells by increasing the
Bax: Bcl-xL ratio through concurrent mechanisms dependent and
independent of its mTOR inhibitory activity. Oncogene.
24:3348–3357. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Sucularli C, Senturk S, Ozturk M and Konu
O: Dose- and time-dependent expression patterns of zebrafish
orthologs of selected E2F target genes in response to serum
starvation/replenishment. Mol Biol Rep. 38:4111–4123. 2011.
View Article : Google Scholar
|
|
42
|
Dallas PB, Gottardo NG, Firth MJ, Beesley
AH, Hoffmann K, Terry PA, Freitas JR, Boag JM, Cummings AJ and Kees
UR: Gene expression levels assessed by oligonucleotide microarray
analysis and quantitative real-time RT-PCR-how well do they
correlate? BMC Genomics. 6:592005. View Article : Google Scholar
|
|
43
|
Yamaguchi Y, Passeron T, Hoashi T, Watabe
H, Rouzaud F, Yasumoto K, Hara T, Tohyama C, Katayama I, Miki T and
Hearing VJ: Dickkopf 1 (DKK1) regulates skin pigmentation and
thickness by affecting Wnt/beta-catenin signaling in keratinocytes.
FASEB J. 22:1009–1020. 2008. View Article : Google Scholar
|
|
44
|
Koch PB, Keys DN, Rocheleau T, Aronstein
K, Blackburn M, Carroll SB and ffrench-Constant RH: Regulation of
dopa decarboxylase expression during colour pattern formation in
wild-type and melanic tiger swallowtail butterflies. Development.
125:2303–2313. 1998.PubMed/NCBI
|
|
45
|
Schallreuter KU and Wood JM: The
importance of L-phenylalanine transport and its autocrine turnover
to L-tyrosine for melanogenesis in human epidermal melanocytes.
Biochem Biophys Res Commun. 262:423–428. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Kwak SS, Suk J, Choi JH, Yang S, Kim JW,
Sohn S, Chung JH, Hong YH, Lee DH, Ahn JK, et al: Autophagy
induction by tetrahydrobiopterin deficiency. Autophagy.
7:1323–1334. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Zitserman D, Gupta S, Kruger WD,
Karbowniczek M and Roegiers F: The TSC1/2 complex controls
Drosophila pigmentation through TORC1-dependent regulation of
catecholamine biosynthesis. PLoS One. 7:e487202012. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Wenzlau JM, Garl PJ, Simpson P, Stenmark
KR, West J, Artinger KB, Nemenoff RA and Weiser-Evans MC: Embryonic
growth-associated protein is one subunit of a novel N-terminal
acetyltransferase complex essential for embryonic vascular
development. Circ Res. 98:846–855. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Goldsmith MI, Iovine MK, O'Reilly-Pol T
and Johnson SL: A developmental transition in growth control during
zebrafish caudal fin development. Dev Biol. 296:450–457. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Moriyama Y, Ohata Y, Mori S, Matsukawa S,
Michiue T, Asashima M and Kuroda H: Rapamycin treatment causes
developmental delay, pigmentation defects and gastrointestinal
malformation on Xenopus embryogenesis. Biochem Biophys Res Commun.
404:974–978. 2011. View Article : Google Scholar
|