Introduction
Epilepsy is one of the most common types of
neurological disorder, which is caused by genetic and acquired
factors (1). In terms of the
genetics of idiopathic epilepsy, it is likely that an increasing
number of genes encoding voltage-gated ion channel subunits,
channel-associating proteins, neurotransmitters and neuropeptide
receptors modify these phenomena in various types of idiopathic
human epilepsy and in different animal models (2). Consistent with this hypothesis, it
has been suggested that up to 1,000 genes may be involved in the
pathogenesis and evolution of epilepsy (3). Identifying the mechanisms, with
particular emphasis on candidate genes, is crucial to improve
current understanding of epileptogenicity and epilepsy
susceptibility.
The procedures of gene profiling by genomic,
transcriptomic and proteomic analyses may provide a useful tool in
identifying candidate genes, which are of importance in epilepsy. A
number of genes have been revealed from genome-wide association
studies (GWAS) for focal and generalized types of epilepsy
(4). De novo mutations in
GABA A receptor (GABR)β3 and UDP-N-acetylglucosamine transferase
have previously been reported to be associated with epilepsy
(5). Emerging evidence has
indicated that synaptic transmission genes, including dynamin 1,
also cause epileptic encephalopathies (6). DNA microarrays and proteomes have
been widely applied to analyze key molecular alterations in human
and classical experimental epilepsy. According to previous reports,
the differentially expressed genes represent several cellular
events, including immune response (7), synaptic transmission (6,8),
synaptic plasticity (9) and
receptor processes (10–12). These genome-wide profiles have
assisted in understanding the complex molecular mechanisms of
epilepsy, however, integrative analyses are often capable of
providing more detailed biological insight (13). The integration of bioinformatics
offers promise in revealing the cellular networks of human
epilepsy.
Lists of potential epileptogenesis-associated genes
from GWAS, DNA microarrays and proteomes have become available and
there is an increasing need to extract functional information from
the high-throughput data. Thus, the present study aimed to
comprehensively analyze the gene expression profile and search for
potential key genes and pathways in epilepsy. As the different
temporal-spatial gene expression patterns are directly related to
the development of epileptic syndromes, the integrative analysis is
crucial to providing novel insights into molecular events and
supports the feasibility of a novel biomarker in epilepsy.
Materials and methods
Data sources
All the human, rat and mouse data obtained in the
present study were from previously published results (http://devicelinked.com/ch/main2.html).
The genes selected in the present study were those identified as
significantly upregulated or downregulated between the epilepsy
group and control group, which was determined as P<0.05,
compared with the control in the previous report. Gene ID
conversions were performed on the collected data using Database to
Database Conversions (bioDBnet; http://biodbnet.abcc.ncifcrf.gov/db/db2db.php)
(14), which was then corrected
and updated by The National Center for Biotechnology Information
(http://www.ncbi.nlm.nih.gov/). The
duplicate genes identified in the same organism were excluded in
the final database. Venn diagrams (http://bioinfogp.cnb.csic.es/tools/venny/) were used
for comparing the gene lists and identifying the associations
between the collected sets.
Chromosome mapping and chromosome
enrichment analysis
The chromosomal locus of each gene was determined
using the Database to Database Conversions at bioDBnet. In
addition, a search was performed on the Entrez Gene Database to
correct the results. A hypergeometric test was used to determine
whether the candidate genes for epilepsy were significantly
enriched in specific chromosomes. The P-value of the test was
defined as follows:
In the above equation, N represents the number of all the genes
annotated with chromosome location information, K represents the
number of epilepsy candidate genes collected. M represents the
number of genes in a specific chromosome and X represents the
number of candidate genes in the same chromosome. The present study
determined the enrichment of the candidate genes in each chromosome
for each species. P<0.05, was considered to indicate a candidate
gene, which was significantly enriched in a specific chromosome.
The data source for the number of genes on each chromosome was the
Ensembl genome browser release 68, July 2012 ().
GO and KEGG pathway enrichment
The Database for Annotation, Visualization and
Integrated Discovery website (DAVID; http://david.abcc.ncifcrf.gov/ (15,16)
was used to identify the significant GO and KEGG pathways in the
differentially expressed gene lists. Multiple corrections were
performed using Fisher's exact method, and DAVID provided a
Benjamini-Hochberg false discovery rate-adjusted P-value.
Protein network analysis
The web-based expression analysis program, Cytoscape
(http://www.cytoscape.org/), was used for
network analysis, to describe the functional associations between
genes or proteins using all human genes and the homologous genes
from rat and mouse candidate gene lists.
RT-qPCR
Three pairs of brain tissue samples from 3 patients
(Table I) with epilepsy were
obtained from Wuhan General Hospital of Guangzhou Military Command
(Wuhan, China), following approval of the Ethics Committee of
South-Central University for Nationalities (Wuhan, China). Written
informed consent was obtained from patients. On the day of RNA
isolation, the tissues were disrupted and homogenzed using TRIzol
reagent (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA,
USA) and used as a template for reserve transcription. A 25
μl reaction was set up containing 1 μg RNA, 0.5
μg Oligo(dT), 5 μl 5X M-MLV buffer, 0.5 mM dNTP each,
25 units of RNasin Ribonuclease Inhibitor and 200 units M-MLV
reverse transcriptase (all purchased from Promega, Madison, WI,
USA). First-strand synthesis reaction was performed at 37°C for 1
h. The qPCR primers used are presented in Table II, and the template samples used
are listed in Table I. A total of
1 μl cDNA was added to a 20 μl reaction volume,
resulting in a final concentration of 400 nM forward primer, 400 nM
reverse primer and 1X SYBR Premix Ex Taq II (Takara Bio, Inc.,
Tokyo, Japan). The qPCR conditions were optimized in an Mx3005P
system (Applied Biosystems; Thermo Fisher Scientific, Inc.). qPCR
was performed at 95°C for 5 min, followed by 40 cycles of 95°C for
30 sec and 61°C for 1 min. The relative expression values were
normalized to the housekeeping gene, GAPDH, and calculated using
the ΔΔCq method (17).
 | Table ISources of epilepsy samples used in
reverse trancsription-quantitative polymerase chain reaction
analysis. |
Table I
Sources of epilepsy samples used in
reverse trancsription-quantitative polymerase chain reaction
analysis.
| Patient | Age (years) | Gender | Epileptic syndrome
diagnosis |
|---|
| 1 | 29 | Male | Refractory
epilepsy |
| 2 | 18 | Male | Refractory
epilepsy |
| 3 | 37 | Male | Refractory
epilepsy |
 | Table IIPrimer sequences for reveres
transcription-quantitative PCR analysis. |
Table II
Primer sequences for reveres
transcription-quantitative PCR analysis.
| Primer | Sequence (5′–3′) | Length (bp) |
|---|
| GRB2s |
AATGAAGCCGTCTTTTCCATT | 21 |
| GRB2a |
ACGAGCTGAGCTTCAAAAGG | 20 |
| APPs |
CCACAGAACATGGCAATCTG | 20 |
| APPa |
TTTGGCACTGCTCCTGCT | 18 |
| TGF-β1s |
CTTCCAGCCGAGGTCCTT | 18 |
| TGF-β1a |
CCCTGGACACCAACTATTGC | 20 |
| VEGFs |
AGCTGCGCTGATAGACATCC | 20 |
| VEGFa |
CTACCTCCACCATGCCAAGT | 20 |
| CDKN1As |
CATGGGTTCTGACGGACAT | 19 |
| CDKN1Aa |
AGTCAGTTCCTTGTGGAGCC | 20 |
| HPRT1s |
GTTATGGCGACCCGCAG | 17 |
| HPRT1a |
ACCCTTTCCAAATCCTCAGC | 20 |
| GAPDHs |
AAGGTGAAGGTCGGAGTCAA | 20 |
| GAPDHa |
AATGAAGGGGTCATTGATGG | 20 |
Results
Gene expression profiling and chromosome
mapping
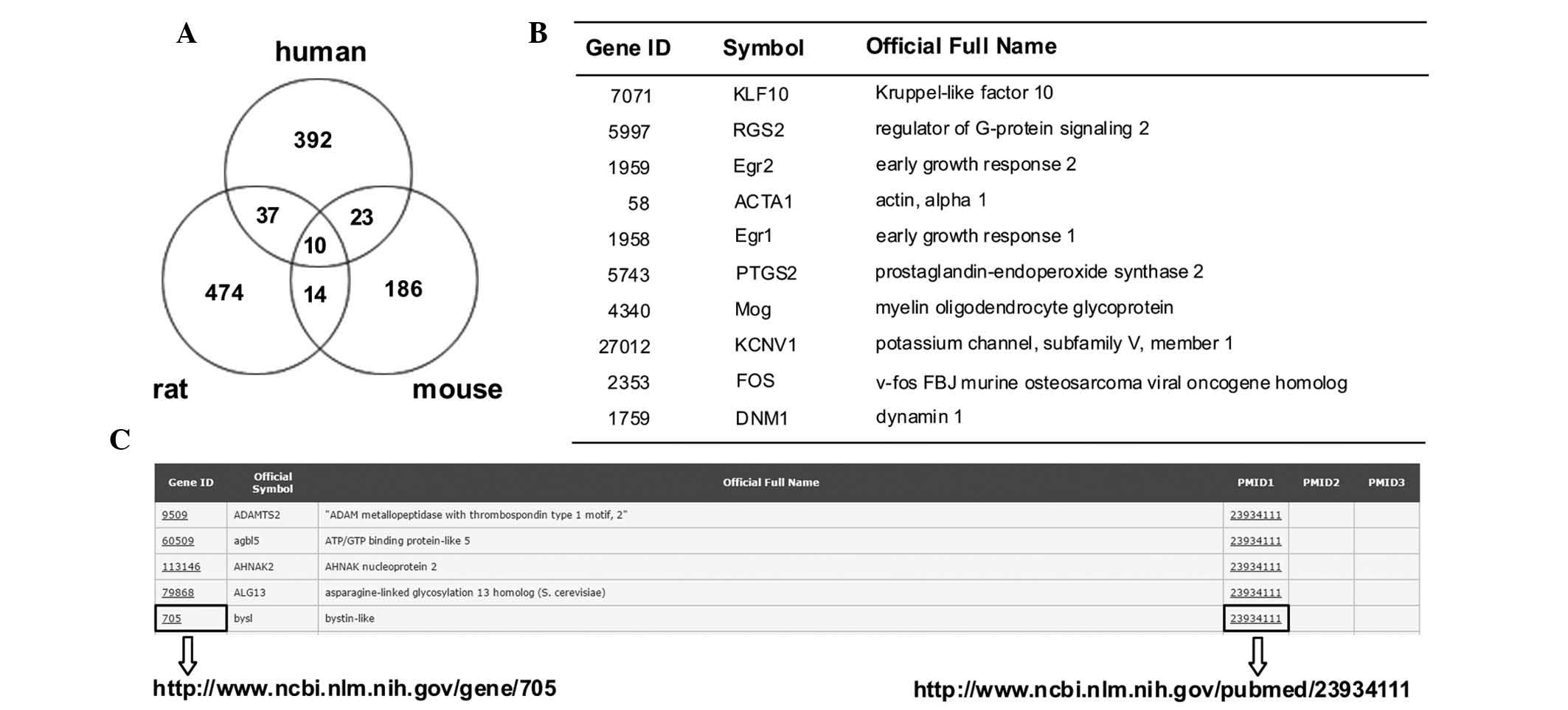
A total number of 1,065 genes were selected, which
showed differential expression in the human epilepsy and animal
models (Fig. 1A and B). The
Candidate Epilepsy Gene Database (http://devicelinked.com/ch/main2.html) is a website
built by the authors and was used to enable rapid querying of
human, rat and mouse genes (Fig.
1C) and to download the candidate genes. The database presents
gene ID, symbol and name, and provides the PubMed ID. The database
is updated when new data is released.
To determine whether there was a chromosome
preference for epilepsy, the differentially expressed genes were
further mapped onto chromosomes. Based on chromosome locations, the
epilepsy genes in humans were enriched on chromosome 1
(hypergeometric test, P=0.048), chromosome 5 (P=0.035) and
chromosome 11 (P=0.045), as shown in Fig. 2A. The genes were enriched on
chromosome 1 (P=0.001), chromosome 20 (P=0.004) and the
mitochondrial chromosome (P=0.003) in the rat (Fig. 2B), and were predominantly located
on chromosome 3 (P=0.008) and 17 (P=0.002) in the mouse (Fig. 2C).
Biological relevance and signaling
pathways associated with epileptogenesis
The GO terms showing the highest level of
significant enrichment are indicative of a bias in the type of
functional classification. The statistically significant biological
processes (BPs) with the highest levels of enrichment were response
to organic substance, intracellular signaling cascade and
neurological system process (adjusted P<0.01). The cellular
component ontology-term enrichment suggested that the important
processes in the development of epileptogenesis occurred in the
cytosol. The prevalent molecular functions were gated channel
activity and structural constituent of ribosome (Table III). Using the KEGG database for
analysis, it was found that the most significant pathways were the
hsa04010 mitogen-activated protein kinase (MAPK) signaling pathway
(31 counts; adjusted P=3.47E-07) and hsa03010 Ribosome (18 counts;
adjusted P=5.43E-07). The rat and mouse data were similar to the
human data obtained, and these can be downloaded from the website
written by the authors (http://devicelinked.com/ch/main2.html).
 | Table IIIGO analysis of epilepsy candidate
genes in humans. |
Table III
GO analysis of epilepsy candidate
genes in humans.
A, GOTERM_BP_FAT
|
|---|
| Term | Count | % | P-value | Adjusted
P-value |
|---|
| GO:0010033 response
to organic substance | 60 | 13.0719 | 1.60E-13 | 1.99E-10 |
| GO:0007242
intracellular signaling cascade | 60 | 13.0719 | 8.54E-05 | 3.71E-03 |
| GO:0050877
neurological system process | 57 | 12.4183 | 1.95E-04 | 6.88E-03 |
| GO:0007267
cell-cell signaling | 50 | 10.8932 | 2.52E-11 | 1.25E-08 |
| GO:0042981
regulation of apoptosis | 48 | 10.4575 | 2.14E-06 | 1.66E-04 |
| GO:0043067
regulation of programmed cell death | 48 | 10.4575 | 2.79E-06 | 2.09E-04 |
| GO:0042592
homeostatic process | 48 | 10.4575 | 3.08E-06 | 2.12E-04 |
| GO:0019226
transmission of nerve impulse | 45 | 9.8039 | 4.37E-06 | 2.71E-04 |
| GO:0009719 response
to endogenous stimulus | 43 | 9.3681 | 2.18E-15 | 5.51E-12 |
| GO:0007268 synaptic
transmission | 36 | 7.8431 | 5.05E-09 | 1.79E-06 |
B, GOTERM_CC_FAT
|
|---|
| Term | Count | % | P-value | Adjusted
P-value |
|---|
| GO:0005829
cytosol | 77 | 16.7755 | 1.51E-10 | 1.76E-08 |
| GO:0044421
extracellular region part | 49 | 10.6753 | 2.44E-05 | 4.72E-04 |
| GO:0031982
vesicle | 42 | 9.1503 | 8.05E-07 | 2.81E-05 |
| GO:0031988
membrane-bounded vesicle | 40 | 8.7145 | 8.12E-08 | 4.05E-06 |
| GO:0031410
cytoplasmic vesicle | 40 | 8.7145 | 1.84E-06 | 4.94E-05 |
| GO:0005615
extracellular space | 40 | 8.7145 | 8.69E-06 | 1.90E-04 |
| GO:0042995 cell
projection | 39 | 8.4967 | 2.98E-05 | 5.20E-04 |
| GO:0016023
cytoplasmic membrane-bounded vesicle | 38 | 8.2788 | 3.01E-07 | 1.17E-05 |
| GO:0043005 neuron
projection | 33 | 7.1895 | 8.95E-10 | 7.81E-08 |
| GO:0045202
synapse | 30 | 6.5359 | 1.16E-07 | 5.05E-06 |
C. GOTERM_MF_FAT
|
|---|
| Term | Count | % | P-value | Adjusted
P-value |
|---|
| GO:0022836 gated
channel activity | 23 | 5.0108 | 4.75E-05 | 7.73E-03 |
| GO:0003735
structural constituent of ribosome | 19 | 4.1394 | 8.50E-07 | 5.56E-04 |
| GO:0022843
voltage-gated cation channel activity | 17 | 3.7037 | 2.82E-06 | 9.21E-04 |
| GO:0022843
voltage-gated cation channel activity | 17 | 3.7037 | 2.82E-06 | 9.21E-04 |
| GO:0005248
voltage-gated sodium channel activity | 6 | 1.3071 | 3.66E-05 | 7.94E-03 |
Top networks during epileptogenesis
In addition to the classical pathways, the present
study aimed to identify the genes, which were likely to be hubs.
Growth factor receptor bound 2 (GRB2) was characterized as an
interconnected node, suggesting it is associated with
epileptogenesis (Fig. 3A). Amyloid
β (A4) precursor protein (APP) was also identified as a hub gene
with a high level of connection (Fig.
3B). Transforming growth factor-β (TGF-β) and vascular
endothelial growth factor (VEGF) were key regulators of the
underlying the protein network (Fig.
3C and D). Cyclin-dependent kinase inhibitor 1 (CDKN1A), also
known as P21, may also exert its role as a central node (Fig. 3E). The networks also indicated that
jun proto-oncogene, tumor necrosis factor, heat shock proteins and
GABRA were also important genes (data not shown).
mRNA levels of GRB2, APP, TGF-β, VEGF and
CDKN1A are upregulated in the cortex of patients with epilepsy
To confirm the hypothesis that the networks
involving GRB2, APP, TGF-β, VEGF and CDKN1A may be important in
human epileptic patients, the present study compared the mRNA
levels of these genes between the epileptic locus, which was
revealed using video-electroencephalography (EEG) recordings, and
in the resection margin, where the incision was made in surgery,
from the same surgically removed cortex sample from patients with
drug-refractory epilepsy (n=3). RT-qPCR analysis revealed that the
mRNA levels of GRB2, APP, TGF-β, VEGF and CDKN1A were all
upregulated in the epileptic cortex samples, compared with the
control (resection margin) samples, in which GAPDH was used as a
reference gene (Fig. 4). The
housekeeping gene, hyperparathyroidism 1, exhibited identical
values in the control and epileptic cortex samples.
Discussion
Several well-characterized models have been
described previously, which represent the complex partial seizures
observed in patients with epilepsy in several ways. Seizures can be
induced by an excitotoxic compound for example kainic acid or
pilocarpine, or by electric stimulation, referred to as kindling
(18,19). Animal models appear to be
particularly informative for assessing the molecular mechanisms
controlling the dynamic processes. Based on the analyses of the
genomic, transcriptional and proteomic data from human patients and
animal models in the present study, specific patterns regarding the
candidate genes were observed, as follows: i) These genes show
chromosomal preference, with a preference to chromosomes 1, 5 and
11 in humans. ii) Epilepsy-associated genes appear to converge on
specific BPs, including response to organic substance,
intracellular signaling cascade and neurological system process. In
addition, the MAPK signaling pathway is involved in multiple
aspects of epileptic seizures. iii) Protein network analysis
suggested that GRB2, APP, TGF-β, VEGF and CDKN1A are key molecules
involved in epileptic seizures, and the expression levels of these
genes are upregulated in epilepsy loci, compared with controls in
the same patient.
Considering the conserved synteny between the human,
rat and mouse genomes, human chromosome 1 has been shown to be
similar to regions of rat chromosome 2 and mouse chromosome 3
(20), on which the epilepsy
candidate genes are all significantly enriched, suggesting that the
orthologous segments on human chromosome 1 may be important in
epileptogenicity. The associated genes also exhibit preference to
human chromosome 5 and its orthologous segment rat chromosome 2.
The gene-rich human chromosome 11 is syntenic with a region of rat
chromosome 1, on which the differentially expressed genes across
rat models are predominantly located. Taken together, the conserved
syntenic clusters of candidate genes on human chromosomes 1, 5 and
11 are likely to be essential for epileptogenesis.
The RT-qPCR analysis of human epilepsy tissue
samples in the present study produced results consistent with those
of previous studies. Specifically, GRB2 (21) and CDKN1A (22) have been reported to be upregulated
in human epilepsy, and APP (23)
and TGF-β (24,25) were found to be upregulated
following epileptic seizures in rat a model, based on DNA
microarrays. Compared with wild-type mice, RT-qPCR has shown that
the mRNA expression of VEGF is 1.44-fold higher in the hippocampus
of VEGF Receptor-2 (Flk-1)-overexpressing mice, characterized by an
elevated threshold for seizure induction (26). Immunostaining has also revealed
that the protein levels of VEGF are increased in neurons and glia
following pilocarpine-induced status epilepticus in rats (20).
In particular, the high proportion of genes from the
GRB2 network may contribute to epileptogenesis or the recovery
process (27,28). The mutations in APP and its
associated genes may lead to toxic accumulation of Aβ protein
fragments, which are important in temporal lobe epilepsy (29). The role of VEGF following seizures
may be either protective or destructive (20). VEGF is important in initiating
changes in the blood-brain barrier (BBB) by vascular remodeling,
and BBB permeability in turn contributes to epileptogenesis
(30). However, VEGF has also been
shown to potentially protect vulnerable cells from the damage
associated with seizures (20).
Consistent with previous reports describing the role of the TGF-β
pathway in the development of neocortical epileptogenesis, TGF-β
may initiate and maintain cellular alterations as a putative
signaling cascade (31,32). Until now, there has been no report
on the exact role of CDKN1A in epilepsy. A subset of these genes
may serve as novel biomarkers to improve the current diagnosis of
epilepsy. The large-scale expression profiling performed in the
present study may provide clues on the epigenetic mechanisms
through the key modules, described above.
The results of the present study offer insights into
the pathogenesis of epilepsy, and a number of the candidate genes
detected may be important in epilepsy. However, further
investigations are required to determine which genes initiate the
occurrence of epilepsy and to determine the exact roles.
Acknowledgments
This study was supported by the National Natural
Science foundation of China (grant nos. 30972848, 31500996 and
81271234) and the Special Funds of Basic research operating
expenses for universities of China (grant nos. CZY14016, CZZ11009,
CZW14022 and CZW14058).
Abbreviations:
|
GWAS
|
genome-wide association studies
|
|
RT-qPCR
|
reverse transcription-quantitative
polymerase chain reaction
|
|
GRB2
|
growth factor receptor bound 2
|
|
APP
|
amyloid β (A4) precursor protein
|
|
TGF-β
|
transforming growth factor β
|
|
VEGF
|
vascular endothelial growth factor
|
|
CDKN1A
|
cyclin-dependent kinase inhibitor
1
|
References
|
1
|
Berg AT, Berkovic SF, Brodie MJ,
Buchhalter J, Cross JH, van Emde Boas W, Engel J, French J, Glauser
TA, Mathern GW, et al: Revised terminology and concepts for
organization of seizures and epilepsies: Report of the ILAE
commission on classification and terminology, 2005–2009. Epilepsia.
51:676–685. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Meldrum BS and Rogawski MA: Molecular
targets for antiepileptic drug development. Neurotherapeutics.
4:18–61. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Frankel WN: Detecting genes in new and old
mouse models for epilepsy: A prospectus through the magnifying
glass. Epilepsy Res. 36:97–110. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Jensen FE: Epilepsy in 2013: Progress
across the spectrum of epilepsy research. Nat Rev Neurol. 10:63–64.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Epi4K Consortium; Epilepsy Phenome/Genome
Project; Allen AS, Berkovic SF, Cossette P, Delanty N, Dlugos D,
Eichler EE, Epstein MP, Glauser T, et al: De novo mutations in
epileptic encephalopathies. Nature. 501:217–221. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
EuroEPINOMICS-RES Consortium, Epilepsy
Phenome/Genome Project, Epi4K Consortium: De novo mutations in
synaptic transmission genes including DNM1 cause epileptic
encephalopathies. Am J Hum Genet. 95:360–370. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Vezzani A, French J, Bartfai T and Baram
TZ: The role of inflammation in epilepsy. Nat Rev Neurol. 7:31–40.
2011. View Article : Google Scholar
|
|
8
|
Fukata Y, Lovero KL, Iwanaga T, Watanabe
A, Yokoi N, Tabuchi K, Shigemoto R, Nicoll RA and Fukata M:
Disruption of LGI1-linked synaptic complex causes abnormal synaptic
transmission and epilepsy. Proc Natl Acad Sci. 107:3799–3804. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Vissel B, Royle G, Christie B, Schiffer
HH, Ghetti A, Tritto T, Perez-Otano I, Radcliffe RA, Seamans J,
Sejnowski T, et al: The role of RNA editing of kainate receptors in
synaptic plasticity and seizures. Neuron. 29:217–227. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Gorter JA, van Vliet EA, Aronica E, Breit
T, Rauwerda H, Lopes da Silva FH and Wadman WJ: Potential new
antiepileptogenic targets indicated by microarray analysis in a rat
model for temporal lobe epilepsy. J Neurosci. 26:11083–11110. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tecott LH, Sun LM, Akana SF, Strack AM,
Lowenstein DH, Dallman MF and Julius D: Eating disorder and
epilepsy in mice lacking 5-HT2c serotonin receptors. Nature.
374:542–546. 1995. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Brooks-Kayal AR, Shumate MD, Jin H,
Rikhter TY and Coulter DA: Selective changes in single cell GABAA
receptor subunit expression and function in temporal lobe epilepsy.
Nat Med. 4:1166–1172. 1998. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lukasiuk K and Pitkänen A: Gene and
protein expression in experimental status epilepticus. Epilepsia.
48(Suppl 8): S28–S32. 2007. View Article : Google Scholar
|
|
14
|
Mudunuri U, Che A, Yi M and Stephens RM:
bioDBnet: The biological database network. Bioinformatics.
25:555–556. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Huang da W, Sherman BT and Lempicki RA:
Bioinformatics enrichment tools: Paths toward the comprehensive
functional analysis of large gene lists. Nucleic Acids Res.
37:1–13. 2009. View Article : Google Scholar
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2−ΔΔCt method. Methods. 25:402–408. 2001. View Article : Google Scholar
|
|
18
|
Sutula TP: Secondary epileptogenesis,
kindling and intractable epilepsy: A reappraisal from the
perspective of neural plasticity. Int Rev Neurobiol. 45:355–386.
2001. View Article : Google Scholar
|
|
19
|
Löscher W: Critical review of current
animal models of seizures and epilepsy used in the discovery and
development of new antiepileptic drugs. Seizure. 20:359–368. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Gibbs RA, Weinstock GM, Metzker ML, Muzny
DM, Sodergren EJ, Scherer S, Scott G, Steffen D, Worley KC, Burch
PE, et al: Genome sequence of the Brown Norway rat yields insights
into mammalian evolution. Nature. 428:493–521. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Arion D, Sabatini M, Unger T, Pastor J,
Alonso-Nanclares L, Ballesteros-Yáñez I, García Sola R, Muñoz A,
Mirnics K and DeFelipe J: Correlation of transcriptome profile with
electrical activity in temporal lobe epilepsy. Neurobiol Dis.
22:374–387. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Beaumont TL, Yao B, Shah A, Kapatos G and
Loeb JA: Layer-specific CREB target gene induction in human
neocortical epilepsy. J Neurosci. 32:14389–14401. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Winden KD, Karsten SL, Bragin A, Kudo LC,
Gehman L, Ruidera J, Geschwind DH and Engel J Jr: A systems level,
functional genomics analysis of chronic epilepsy. PloS One.
6:e207632011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Okamoto OK, Janjoppi L, Bonone FM, Pansani
AP, da Silva AV, Scorza FA and Cavalheiro EA: Whole transcriptome
analysis of the hippocampus: Toward a molecular portrait of
epileptogenesis. BMC Genomics. 11:2302010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Christensen KV, Leffers H, Watson WP,
Sánchez C, Kallunki P and Egebjerg J: Levetiracetam attenuates
hippocampal expression of synaptic plasticity-related immediate
early and late response genes in amygdala-kindled rats. BMC
Neurosci. 11:92010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Nikitidou L, Kanter-Schlifke I, Dhondt J,
Carmeliet P, Lambrechts D and Kokaia M: VEGF receptor-2 (Flk-1)
overexpression in mice counteracts focal epileptic seizures. PloS
One. 7:e405352012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Pitkänen A and Lukasiuk K: Molecular and
cellular basis of epileptogenesis in symptomatic epilepsy. Epilepsy
Behav. 14(Suppl 1): S16–S25. 2009. View Article : Google Scholar
|
|
28
|
Newton SS, Collier EF, Bennett AH, Russell
DS and Duman RS: Regulation of growth factor receptor bound 2 by
electroconvulsive seizure. Brain Res. 129:185–188. 2004. View Article : Google Scholar
|
|
29
|
Noebels J: A perfect storm: Converging
paths of epilepsy and Alzheimer's dementia intersect in the
hippocampal formation. Epilepsia. 52(Suppl 1): S39–S46. 2011.
View Article : Google Scholar
|
|
30
|
Morin-Brureau M, Lebrun A, Rousset MC,
Fagni L, Bockaert J, de Bock F and Lerner-Natoli M: Epileptiform
activity induces vascular remodeling and zonula occludens 1
downregulation in organotypic hippocampal cultures: Role of VEGF
signaling pathways. J Neurosci. 31:10677–10688. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Friedman A, Kaufer D and Heinemann U:
Blood-brain barrier breakdown-inducing astrocytic transformation:
Novel targets for the prevention of epilepsy. Epilepsy Res.
85:142–149. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Friedman A: Blood-brain barrier
dysfunction, status epilepticus, seizures and epilepsy: A puzzle of
a chicken and egg? Epilepsia. 52(Suppl 8): S19–S20. 2011.
View Article : Google Scholar
|