Introduction
Thyroid cancer is the most common endocrine
malignancy and its incidence is increasing (1). The molecular mechanisms underlying
the development and progression of thyroid have remained to be
fully elucidated. Genetic and epigenetic alterations in the major
signaling pathways are central to these mechanisms and genes
affected represent novel molecular markers and therapeutic targets,
providing a basis for further research and clinical development of
treatment strategies for thyroid cancer (2,3).
SHARP1, also known as basic helix-loop-helix family
member e41 (BHLHE41), BHLHB3 or Dec2, is a basic helix-loop-helix
transcription factor that has complex roles in cellular
differentiation, apoptosis and tumor progression (4–6). It
is expressed at high levels in the brain and skeletal muscle, at
moderate levels in the pancreas and heart, and at low levels in the
lung and placenta, while it is barely detectable in the liver and
kidneys (7,8). SHARP1 has also been reported to be
involved in mutant p53-mediated metastasis (9,10).
Montagner et al (11)
identified a significant association between HIF activity and
SHARP1 expression in a cohort of triple-negative breast cancer
(TNBC) patients. Furthermore, SHARP1 suppresses breast cancer
metastasis and negatively regulates vascular endothelial growth
factor (VEGF) expression (11,12).
However, whether and how SHARP1 contributes to the development and
progression of thyroid cancer has remained elusive.
Hypoxia in tissues is responsible for cell
metabolism reprogramming, thus increasing cell proliferation,
transformation and cancer progression (13). Hypoxia-inducible factor-1 (HIF-1)
is a transcriptional activator and helps cells adapt to hypoxia.
The transcriptional activity of HIF-1 is regulated by HIF-1α, which
activates multiple target genes involved in cancer biology,
inducing cell proliferation, survival, apoptosis and angiogenesis
(14–16). A number of studies showed that a
reduction in HIF-1α protein was a consequence of
proteasome-dependent degradation by SHARP1 (17,18).
This led to the hypothesis that SHARP1 acts as a global inhibitor
of HIF-1α activity. However, the influences of SHARP1 on HIF-1α
expression in thyroid cancer cells have not been assessed to date,
to the best of our knowledge.
The present study revealed that SHARP1 was
downregulated in certain thyroid cancer cell lines as well as in
thyroid cancer tissues. Overexpression of SHARP1 inhibited the
viability, migration and invasion of thyroid cancer cells, while
reducing the protein levels of HIF-1α in parallel. These results
indicated that SHARP1 functions as a tumor suppressor in thyroid
cancer, possibly via reduction of HIF-1α levels, and may therefore
represent a potential therapeutic target for the treatment of
thyroid cancer.
Materials and methods
Cell culture
All culture media were supplemented with 10% fetal
bovine serum (FBS), 100 U/ml penicillin G and 100 mg/ml
streptomycin (all from Thermo Fisher Scientific, Inc., Waltham, MA,
USA). The thyroid cancer cell lines TT, TPC-1, FTC-133 and ARO as
well as the Nthy-ori 3-1 normal thyroid cell line were obtained
from the Cell Bank of Academia Sinica (Shanghai, China). Cells were
maintained in Dulbecco's Modified Eagle's Medium (DMEM) in a
humidified incubator containing 5% CO2 in air at
37°C.
Patient samples
A total of 30 pairs of thyroid cancer tissues and
matched normal tissues were obtained from patients (n=12 women and
n=18 men; age, 14–91 years; median age, 53 years) who underwent
surgery between October 2012 to February 2015. at Yangpu Hospital
(Shanghai Tongji University School of Medicine, Shanghai, China).
The normal tissues were resected within ≥5 cm of the tumor margin
during surgery. No patients had received radiotherapy or
chemotherapy. The study was approved by the Ethics Review Committee
of the institutional review board of Yangpu Hospital (Shanghai
Tongji University School of Medicine, Shanghai, China) and written
informed consent was obtained from every patient. Tissues were
centrifuged at 400 × g at 25°C for 20 sec for homogenization and
immediately placed on ice, and then centrifuged at 400 × g at 4°C
for 10 min and stored at 80°C prior to experiments.
Reverse-transcription quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted from thyroid cancer cells
with TRIzol reagent (Thermo Fisher Scientific, Inc.) as previously
described (19) and stored at
−80°C. Complementary DNA (cDNA) was synthesized using a cDNA
synthesis kit (Thermo Fisher Scientific, Inc.). The DyNAmo Flash
SYBR Green qPCR kit (Finnzymes Oy, Espoo, Finland) was used for PCR
amplification according to the manufacturer's instructions to
determine the mRNA levels of SHARP1 and HIF-1α genes. The primer
sequences (sense/antisense) used were as follows: SHARP1, 5′-GAC
CAA CTG CTT CAC ACT TTC-3′ and 5′-GCT GTT CGT TTC CTC TGT TTC-3′;
HIF-1α, 5′-TCG GCG AAG TAA AGA ATC-3′ and 5′-TTC CTC ACA CGC AAA
TAG-3′; glyceraldehyde-3-phosphate dehydrogenase (GAPDH): 5′-CAC
CCA CTC CTC CAC CTTTG-3′ and 5′-CCA CCA CCC TGT TGC TGTAG-3′ (all
obtained from Sangon Biotech Co., Ltd., Shanghai, China). The PCR
reaction mixture contained 12.5 μl DyNAmo Flash SYBR Green
qPCR mix (Thermo Fisher Scientific, Inc.), 0.5 μl
forward/reverse primers, 9.5 μl ddH2O and 2
μl cDNA. The PCR cycling conditions were as follows: 95°C
for 10 min, followed by 40 cycles at 95°C for 15 sec and 60°C for
45 sec, and a final extension step of 95°C for 15 sec, 60°C for 1
min, 95°C for 15 sec and 60°C for 15 sec. Data collection was
performed using an ABI 7500 (Thermo Fisher Scientific, Inc.) and
relative quantification of gene expression was performed using the
2−ΔΔCq method (20).
Relative quantification was performed by normalizing the signals of
different genes to the GAPDH signal.
Western blot analysis
Total protein was isolated from thyroid cancer cell
lines with radioimmunoprecipitation buffer (Wuhan Amyjet Scientific
Co., Ltd., Wuhan, China). and the protein concentration was
determined using a BCA protein assay kit (Pierce Biotechnology,
Inc., Rockford, IL, USA). Total protein (30 μg) was
separated using 10–15% sodium dodecyl sulfate-polyacrylamide gel
electrophoresis (Wuhan Amyjet Scientific Co., Ltd.) and transferred
to polyvinylidene fluoride membranes (Sigma-Aldrich, St. Louis, MO,
USA), followed by blocking in non-fat milk overnight at 4°C. The
membrane was first incubated with antibodies against SHARP1 (mouse
polyclonal; 1:1,000; cat. no., ab57739; Abcam, Cambridge, MA, USA),
HIF-1α (mouse polyclonal; 1:1,000; cat. no., ab113642; Abcam) and
GAPDH (rabbit monoclonal; 1:1,000; cat. no. #5174; Cell Signaling
Technology, Inc., Danvers, MA, USA) for 2 h at 25°C. The membranes
were then incubated with horseradish peroxidase conjugated goat
anti-rabbit IgG (cat. no. A0208; 1:1,000; Beyotime Institute of
Biotechnology, Haimen, China) and goat anti mouse IgG (cat. no.
A0216; 1:1,000; Beyotime Institute of Biotechnology) secondary
antibodies for 1 h at 37°C, and washed three times with
Tris-buffered saline with Tween-20 (Amresco, Solon, OH, USA).
SHARP1 and HIF-1α, and then with anti-GAPDH antibody as a loading
control. The membranes were then probed with secondary antibodies
labeled with horseradish peroxidase and signal intensity was
determined using Image J software 1.46 (National Institutes of
Health, Bethesda, MD, USA).
Antibodies of SHARP1 and HIF-1α were purchased from
Abcam (Cambridge, MA, USA) and GAPDH (Cell Signaling Technology,
Inc., Danvers, MA, USA). All primary antibodies were used at a
1:1,000 dilution.
Overexpression vector construction and
transfection
To construct the SHARP1 overexpression vector, a
sequence was designed by Sangon Biotech Co., Ltd. and inserted into
the pLenti6/V5-DEST vector (Thermo Fisher Scientific, Inc.). The
overexpression vectors were transfected into TT and TPC-1 cells
using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific,
Inc.) according to the manufacturer's instructions. An empty vector
was used as a negative control (NC) and the thyroid cancer cells
were analyzed 48 h after transfection.
Transfection of HIF-1α small interfering
(si)RNA
siRNA specific to HIF-1α (5′-TCGGCGAAGTAAAGAATC-3′)
was obtained from Genesil Biotechnology (Wuhan, China). The TT and
TPC 1 cells were plated onto 96-well plates at a density of
2×103 cells/well and were transfected with siRNA (40 nM)
using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific,
Inc.), according to the manufacturer's protocol. A non-specific
scramble siRNA sequence (Genesil Biotechnology) was used as a
negative control. Cells were analyzed 48 h after transfection.
Cell viability assay
A Cell Counting Kit (CCK)-8 assay (Beyotime
Institute of Biotechnology, Inc., Haimen, China) was used to
determine the cell viability according to the manufacturer's
instructions. Following transfection with siHIF-1α for 48 h, TT and
TPC-1 cells were plated onto 96-well plates at a density of
2×103 cells/well. After incubation for 0, 12, 24, 48 or
72 h, 10 μl CCK-8 reagent was added to each well, followed
by incubation at 37°C for 1 h. The absorbance was measured using a
microplate reader (Bio-Rad Laboratories, Inc., Hercules, CA, USA)
at 450 nm and analyzed using Microplate manager 6 software (Bio-Rad
Laboratories, Inc.).
Migration and invasion assays
The migratory and invasive capacity of the cells
following transfection with SHARP1 overexpression vector or HIF-1α
silencing were examined using a Transwell culture system comprised
of Transwell inserts (5 μm; Corning, Corning, NY, USA). with
membranes coated with or without Matrigel (2.5 mg/ml; BD
Biosciences, Franklin Lakes, NJ, USA). Following transfection with
siHIF-1α for 48 h, suspensions of 105 TT and TPC-1
cells/ml in DMEM with 1% FBS were prepared and cells were seeded
into the upper wells of optionally pre-coated Transwells at
5×104 cells per well. The lower wells of the Transwells
contained DMEM with 10% FBS. After 48 h of incubation, cells that
had transgressed through the membrane were washed with
phosphate-buffered saline (PBS), fixed in 4% paraformaldehyde and
stained by 0.5% crystal violet. Images of the lower sides of the
membranes were captured and cells counted under a microscope
(CX41RF; Olympus Corporation, Tokyo, Japan).
Statistical analysis
Values are expressed as the mean ± standard
deviation. The paired, two-tailed Student's t-test was used to
analyze the significance of differences between groups. Statistical
analysis was performed using GraphPad Prism 5 software (GraphPad
Software, Inc., La Jolla, CA, USA). P<0.01 was considered to
indicate a statistically significant difference.
Results
Expression of SHARP1 in thyroid cancer
cell lines and tissues
In order to elucidate the role of SHARP1 in thyroid
cancer, its expression levels were initially examined in the
thyroid cancer cell lines TT, TPC-1, FTC-133 and ARO, and compared
with those in the Nthy-ori 3-1 normal thyroid cell line at the mRNA
and protein level. The results indicated that SHARP1 expression was
in TT and TPC-1 cells was significantly decreased in comparison to
that in Nthy-ori 3-1 cells (P<0.01) (Fig. 1A and B). Therefore, subsequent
experiments were performed on TT and TPC-1 cells. Furthermore, the
expression of SHARP1 was significantly downregulated in thyroid
cancer tissues compared to their matched normal tissues (P<0.01)
(Fig. 1C). These results suggested
that SHARP1 may act as a tumor suppressor in thyroid cancer.
SHARP1 suppresses the viability of TT and
TPC-1 cells
In order to explore the biological significance of
SHARP1 in the genesis of thyroid cancer, TT and TPC-1 cells were
stably transfected with SHARP1 overexpression vector. The efficacy
of transfection was examined by RT-qPCR and western blotting
(Fig. 2A and B). The mRNA
expression of SHARP1 was increased by 3.75 and 2.35 fold in TT and
TPC 1 cells, respectively. The protein expression of SHARP1 was
increased by 1.34- and 1.14-fold in TT and TPC-1 cells,
respectively (P<0.01 vs. NC) (Fig.
2B). Next, the effects of SHARP1 on the proliferation/viability
of TT and TPC-1 cells were assessed. At 72 h after transfection
with SHARP1 overexpression vector, the number of viable TT and
TPC-1 cells was reduced by ~43 and ~42%, respectively (P<0.01
vs. NC) (Fig. 2C and D). These
results suggested that SHARP1 may function as a tumor suppressor in
TT and TPC-1 cells through inhibition of cell
proliferation/viability.
SHARP1 suppresses the migration and
invasion of TT and TPC-1 cells
The migratory capacities of TT and TPC-1 cells with
stably overexpressing SHARP1 were examined using Transwell assays
over 48 h. The number of migratory TT and TPC-1 cells with forced
SHARP1 overexpression decreased by 61.4 and 53.6% compared with the
NC groups, respectively (P<0.01) (Fig. 3A). Furthermore, Transwell invasion
assays with Matrigel-coated membranes showed that SHARP1
overexpression decreased the invasive capacity of TT and TPC-1
cells by 73.4 and 70.1% compared with the NC groups (P<0.01)
(Fig. 3B). These results suggested
that SHARP1 may function as a tumor suppressor in TT and TPC-1
cells through inhibition of cell migration and invasion.
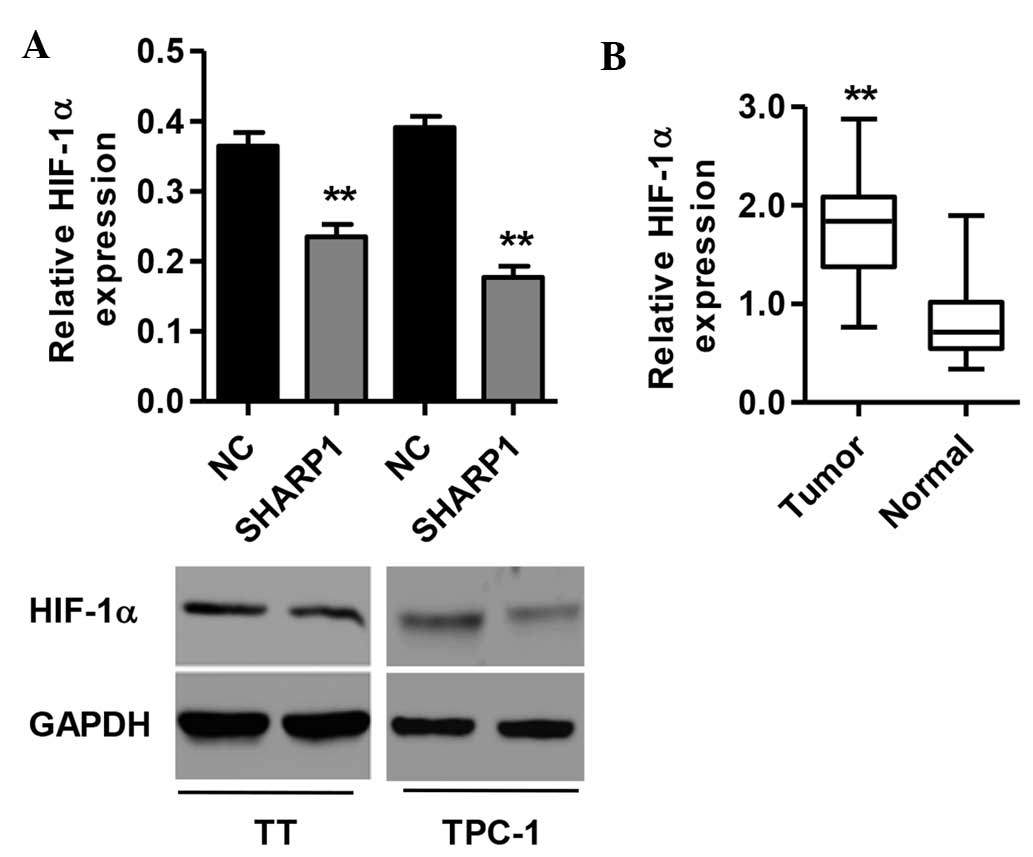
SHARP1 reduces HIF-1α levels in TT and
TPC-1 cells
To elucidate the mechanisms by which SHARP1 experts
its effects, the influence of SHARP1 expression on HIF-1α levels
was assessed. At the mRNA and protein level, HIF-1α was
significantly decreased in SHARP1-overexpressing TT and TPC-1 cells
(P<0.01 vs. NC) (Fig. 4A). The
association between SHARP1 and HIF-1α was further investigated by
analyzing the relative protein levels of HIF-1α in the 30 paired
tumor and normal tissues from thyroid cancer patients, showing that
HIF-1α was significantly increased in tumor tissues compared with
normal tissues (P<0.01) (Fig.
4B). These results indicated that SHARP1 may directly or
indirectly regulate the levels of HIF-1α in thyroid cancer.
HIF-1α knockdown reduces the viability,
migration and invasion of TT and TPC-1 cells
As the above results and previous studies indicated
that SHARP1 affects the expression of HIF-1α in thyroid cancer
cells (17,18), the present study examined the
effects of HIF-1α knockdown on the viability, migration and
invasion of TT and TPC-1 cells. The knockdown efficiency of a
lentiviral vector containing shRNA targeting HIF-1α was confirmed
by western blot analysis (P<0.01 vs. NC) (Fig. 5A). HIF-1α silencing significantly
reduced the proliferation/viability and repressed the migratory and
invasive capacities of TT and TPC-1 cells (P<0.01 vs. NC)
(Fig. 5B–D). These findings
suggested that HIF-1α, whose levels were shown to be affected by
SHARP1, is important for the viability, migration and invasion of
TT and TPC-1 cells.
Discussion
Although thyroid cancer is the most common endocrine
tumor, the underlying molecular mechanisms have remained to be
fully elucidated. SHARP1 has been previously reported to be a clock
gene (21,22), a transcriptional repressor
(23) and importantly, a tumor
suppressor in lung cancer (24).
The present study reported a potential role for SHARP1 as a tumor
suppressor in thyroid cancer, which it may exert by decreasing the
levels of HIF-1α. SHARP1 was found to be decreased in tumor tissues
compared with that in normal tissues; furthermore, SHARP1 was
decreased at the mRNA and protein level in TT and TPC-1 cells
compared with that in other thyroid cancer cell lines and the
Nthy-ori 3-1 normal thyroid cell line. By contrast, a previous
study showed that in human breast cancer cells SHARP1 expression
was increased compared with that in normal human breast cells
(25). The expression of SHARP1
was diffuse in different tumors, suggesting regulation of the
protein via a combination of the tumor genotype and the
microenvironment (11). High
expression of SHARP1 in primary breast cancer tumors has been found
to be an indicator of favorable prognosis (26).
In the present study, stable vector-mediated
overexpression of SHARP1 significantly inhibited the
proliferation/viability of TT and TPC-1 cells, which was consistent
with the results of previous studies on endometrial cancer cells
(17) and disseminated tumor cells
in a model of dormant head and neck squamous cell carcinoma
(27). In addition, SHARP1 is
known to suppress invasion, migration and metastasis by inhibiting
HIFs. Loss of SHARP1 led to an increase in MII-cell (TNBC
non-metastatic MCF10Atk1 cells) migration, which was attenuated by
concomitant silencing of HIF-1α (11). To gain insight into the mechanisms
by which SHARP1 inhibits malignant progression, the present study
further assessed the migration and invasion of TT and TPC-1 cells
following SHARP1 overexpression. The results demonstrated that
overexpression of SHARP1 resulted in a significant inhibition of
migration and invasion of TT and TPC-1 cells. Due to its
anti-proliferative and anti-metastatic roles in human thyroid
cancer, SHARP1 may be a potential therapeutic target worth
pursuing.
The results of the present study raise the question
of whether and how SHARP1 affects the levels and expression of
HIF-1α. SHARP1 binds to the HIF-1α sub-unit and directly shuttles
it to the proteasome for degradation under normoxic as well as
hypoxic condition, followed by downregulation of HIF-1-responsive
genes (28). In TNBC cell lines,
overexpression of SHARP1 led to the suppression of the protein
levels and transcriptional activity of HIF-1α under hypoxia
(29,30). However, in the present study, the
mRNA and protein levels of HIF-1α were significantly decreased in
TT and TPC-1 cells with stable expression of SHARP1 under normoxic
conditions, compared with those in the NC group. These results
suggested that SHARP1 is a hypoxia-independent regulator of HIF-1α
levels. Indeed, low SHARP1 expression has been shown to contribute
to high HIF activity and low metastatic capacity. In a previous
study, overexpressed HIF-1α and SHARP1 co-immunoprecipitated in
Cos7 cells, and SHARP1 overexpression repressed HIF-1α-dependent
control of the VEGF-A promoter (31).
In the present study, HIF-1α was identified as a
target of SHARP1 and SHARP1 overexpression was shown to lead to the
downregulation of HIF-1α as well as inhibition of the
proliferation/viability, migration and invasion of TT and TPC-1
cells. Furthermore, knockdown of HIF-1α had similar effects on the
proliferation/viability, migration and invasion of TT and TPC-1
cells. The findings of the present study may therefore indicate
that the tumor suppressor role of SHARP1 may, at least in part, be
mediated via the regulation of HIF-1α expression. It is important
to note that HIF-1α is activated not only by hypoxia within the
tumor but also by oncogenic stimulation (32). Knockdown of HIF-1α has been shown
to reduce the migration and invasion of glioma cells (33,34).
This regulatory role of HIF-1α on migration and invasion have also
been found in colon (35), lung
(36) and gastric cancer (37). These studies supported the notion
that the inhibition of the proliferation/viability, migration and
invasion of thyroid cancer cells may, at least in part, be due to
downregulation of HIF-1α expression, which was demonstrated to be
an effect of SHARP1 overexpression.
In conclusion, the present study showed that SHARP1
was downregulated in certain thyroid cancer cell lines as well as
in thyroid cancer tissues, and that its ectopic expression
inhibited the proliferation/viability, migration and invasion of
the TT and TPC-1 thyroid cell lines. HIF-1α was found to be
overexpressed in thyroid cancer tissues, and SHARP1 overexpression
decreased the levels of HIF-1α in TT and TPC-1 cells under normoxic
conditions. Furthermore, knockdown of HIF-1α inhibited the
proliferation/viability, migration and invasion of TT and TPC-1
cells. These results indicated that SHARP1 may have important roles
in the proliferation of thyroid cancer cells and the development of
metastasis, possibly via decreasing HIF-1α, and that SHARP1 may
represent a potential therapeutic target for the treatment of
thyroid cancer.
Acknowledgments
This study was supported by the Science and
Technology Commission of Shanghai Municipality Fund (no.
14DZ1941804).
References
|
1
|
Kondo T, Ezzat S and Asa SL: Pathogenetic
mechanisms in thyroid follicular-cell neoplasia. Nat Rev Cancer.
6:292–306. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Xing M: Molecular pathogenesis and
mechanisms of thyroid cancer. Nat Rev Cancer. 13:184–199. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Nikiforov YE and Nikiforova MN: Molecular
genetics and diagnosis of thyroid cancer. Nat Rev Endocrinol.
7:569–580. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Rossner MJ, Oster H, Wichert SP, Reinecke
L, Wehr MC, Reinecke J, Eichele G, Taneja R and Nave KA: Disturbed
clockwork resetting in Sharp-1 and Sharp-2 single and double mutant
mice. PloS One. 3:e27622008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yamada K and Miyamoto K: Basic
helix-loop-helix transcription factors, BHLHB2 and BHLHB3; their
gene expressions are regulated by multiple extracellular stimuli.
Front Biosci. 10:3151–3171. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sun H, Ghaffari S and Taneja R:
bHLH-Orange transcription factors in development and cancer. Transl
Oncogenomics. 2:107–120. 2007.PubMed/NCBI
|
|
7
|
Azmi S, Sun H, Ozog A and Taneja R:
mSharp-1/DEC2, a basic helix-loop-helix protein functions as a
transcriptional repressor of E box activity and Stra13 expression.
J Biol Chem. 278:20098–20109. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Fujimoto K, Shen M, Noshiro M, Matsubara
K, Shingu S, Honda K, Yoshida E, Suardita K, Matsuda Y and Kato Y:
Molecular cloning and characterization of DEC2, a new member of
basic helix-loop-helix proteins. Biochem Biophys Res Commun.
280:164–171. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Muller PA, Vousden KH and Norman JC: p53
and its mutants in tumor cell migration and invasion. J Cell Biol.
192:209–218. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sermeus A and Michiels C: Reciprocal
influence of the p53 and the hypoxic pathways. Cell Death Dis.
2:e1642011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Montagner M, Enzo E, Forcato M, Zanconato
F, Parenti A, Rampazzo E, Basso G, Leo G, Rosato A, Bicciato S, et
al: SHARP1 suppresses breast cancer metastasis by promoting
degradation of hypoxia-inducible factors. Nature. 487:380–384.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wu Y, Sato F, Bhawal UK, Kawamoto T,
Fujimoto K, Noshiro M, Seino H, Morohashi S, Kato Y and Kijima H:
BHLH transcription factor DEC2 regulates pro-apoptotic factor Bim
in human oral cancer HSC-3 cells. Biomed Res. 33:75–82. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Cui J, Mao X, Olman V, Hastings P and Xu
Y: Hypoxia and miscoupling between reduced energy efficiency and
signaling to cell proliferation drive cancer to grow increasingly
faster. J Mol Cell Biol. 4:174–176. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Solaini G, Baracca A, Lenaz G and Sgarbi
G: Hypoxia and mitochondrial oxidative metabolism. Biochim Biophys
Acta. 1797:1171–1177. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bruick RK: Oxygen sensing in the hypoxic
response pathway: Regulation of the hypoxia-inducible transcription
factor. Genes Dev. 17:2614–2623. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Semenza GL: Targeting HIF-1 for cancer
therapy. Nat Rev Cancer. 3:721–732. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liao Y, Lu W, Che Q, Yang T, Qiu H, Zhang
H, He X, Wang J, Qiu M, Zou Y, et al: SHARP1 suppresses
angiogenesis of endometrial cancer by decreasing hypoxia-inducible
factor-1α level. PloS One. 9:e999072014. View Article : Google Scholar
|
|
18
|
Li J, Xu Y, Jiao H, Wang W, Mei Z and Chen
G: Sumoylation of hypoxia inducible factor-1α and its significance
in cancer. Sci China Life Sci. 57:657–664. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Payton JE, Grieselhuber NR, Chang LW,
Murakami M, Geiss GK, Link DC, Nagarajan R, Watson MA and Ley TJ:
High throughput digital quantification of mRNA abundance in primary
human acute myeloid leukemia samples. J Clin Invest. 119:1714–1726.
2009. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real time quantitative PCR and
the 2(Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
21
|
Kawamoto T, Noshiro M, Furukawa M, Honda
KK, Nakashima A, Ueshima T, Usui E, Katsura Y, Fujimoto K, Honma S,
et al: Effects of fasting and re-feeding on the expression of Dec1,
Per1 and other clock-related genes. J Biochem. 140:401–408. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Rohleder N, Langer C, Maus C,
Spiwoks-Becker I, Emser A, Engel L and Spessert R: Influence of
photoperiodic history on clock genes and the circadian pacemaker in
the rat retina. Eur J Neurosci. 23:105–111. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Azmi S, Ozog A and Taneja R: Sharp-1/DEC2
inhibits skeletal muscle differentiation through repression of
myogenic transcription factors. J Biol Chem. 279:52643–52652. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Falvella F, Colombo F, Spinola M,
Campiglio M, Pastorino U and Dragani T: BHLHB3: A candidate tumor
suppressor in lung cancer. Oncogene. 27:3761–3764. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wu Y, Sato F, Bhawal UK, Kawamoto T,
Fujimoto K, Noshiro M, Morohashi S, Kato Y and Kijima H: Basic
helix-loop-helix transcription factors DEC1 and DEC2 regulate the
paclitaxel-induced apoptotic pathway of MCF-7 human breast cancer
cells. Int J Mol Med. 27:491–495. 2011.PubMed/NCBI
|
|
26
|
van't Veer LJ, Dai H, Van De Vijver MJ, He
YD, Hart AA, Mao M, Peterse HL, van der Kooy K, Marton MJ, et al:
Gene expression profiling predicts clinical outcome of breast
cancer. Nature. 415:530–536. 2002. View
Article : Google Scholar
|
|
27
|
Bragado P, Estrada Y, Parikh F, Krause S,
Capobianco C, Farina HG, Schewe DM and Aguirre-Ghiso JA: TGF-β2
dictates disseminated tumour cell fate in target organs through
TGF-β-RIII and p38α/β signalling. Nat Cell Biol. 15:1351–1361.
2013. View
Article : Google Scholar : PubMed/NCBI
|
|
28
|
Foster JG, Wong SC and Sharp TV: The
hypoxic tumor micro-environment: Driving the tumorigenesis of
non-small-cell lung cancer. Future Oncol. 10:2659–2674. 2014.
View Article : Google Scholar
|
|
29
|
Bernardi R and Gianni L: Hallmarks of
triple negative breast cancer emerging at last? Cell Res.
24:904–905. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Burgess DJ: Tumour suppressors: At the
SHARP end of metastasis. Nat Rev Cancer. 12:580–581. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Sato F, Bhawal UK, Kawamoto T, et al:
Basic-helix-loop-helix (bHLH) transcription factor DEC2 negatively
regulates vascular endothelial growth factor expression. Genes
Cells. 13:131–144. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Shafee N, Kaluz S, Ru N and Stanbridge EJ:
PI3K/Akt activity has variable cell-specific effects on expression
of HIF target genes, CA9 and VEGF, in human cancer cell lines.
Cancer Lett. 282:109–115. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Méndez O, Zavadil J, Esencay M, et al:
Research Knock down of HIF-1alpha in glioma cells reduces migration
in vitro and invasion in vivo and impairs their ability to form
tumor spheres. Mol Cancer. 9:1332010. View Article : Google Scholar
|
|
34
|
Fujiwara S, Nakagawa K, Harada H, et al:
Silencing hypoxia-inducible factor-1alpha inhibits cell migration
and invasion under hypoxic environment in malignant gliomas. Int J
Oncol. 30:793–802. 2007.PubMed/NCBI
|
|
35
|
Krishnamachary B, Berg-Dixon S, Kelly B,
Agani F, Feldser D, Ferreira G, Iyer N, LaRusch J, Pak B, Taghavi P
and Semenza GL: Regulation of colon carcinoma cell invasion by
hypoxia-inducible factor 1. Cancer Res. 63:1138–1143.
2003.PubMed/NCBI
|
|
36
|
Li Y, Qiu X, Zhang S, Zhang Q and Wang E:
Hypoxia-induced CCR7 expression via HIF-1alpha and HIF-2alpha
correlates with migration and invasion in lung cancer cells. Cancer
Biol Ther. 8:322–330. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wang Y, Li Z, Zhang H, Jin H, Sun L, Dong
H, Xu M, Zhao P, Zhang B, Wang J, et al: HIF-1α and HIF-2α
correlate with migration and invasion in gastric cancer. Cancer
Biol Ther. 10:376–382. 2010. View Article : Google Scholar : PubMed/NCBI
|