Introduction
Hepatocellular carcinoma (HCC) is one of the most
common malignant cancer types worldwide and is accountable for
almost 600,000 mortalities each year worldwide (1); it is also has the second highest
mortality rate amongst all cancer types in China (2). The main risk factors for HCC
development include chronic hepatitis B and C infection, alcohol
abuse and aflatoxin intake (3,4), as
they induce liver cirrhosis, from which 80% of HCCs are derived
(5). Activation of oncogenes and
inactivation of tumor suppressor genes have been identified to be
associated with carcinogenesis and progression of HCC. Various
genes have been identified to be differentially expressed in HCC
tissues compared with paratumor tissues, including HIWI IGF2,
FAT10, SCARA5, DLK1, p53 and ZNF267 (6–12),
which have either oncogenic or tumor suppressive roles, indicating
that HCC is based on complex oncogenic factors.
Besides oncogene activation and deregulation of
apoptosis-associated genes, inactivation of tumor suppressor genes
has also been associated with HCC (13). Evasion of apoptosis and
angiogenesis are typical cancer-associated processes, whose
reversal is an efficient therapeutic strategy for HCC (14) and other tumor types (15). Inhibitors of apoptosis (IAPs) are
characterized by highly conserved baculoviral IAP repeats (16), belonging to a family of endogenous
inhibitors of caspases (17,18).
X-linked IAP (XIAP) prevents the activities of caspase-3, -7 and -9
via directly binding to these caspases (19). Overexpression of XIAP has been
reported in most human cancer types, including HCC, and to be an
independent prognostic factor for HCC patients (20). Inhibition of XIAP induces apoptosis
and inhibits the growth of HCC cells (21), implying that targeting XIAP may be
a promising approach for HCC therapy. XIAP-associated factor
(XAF)-1 specifically inhibits IAP and sensitizes cancer cells to
apoptosis (22), resulting in a
pro-apoptotic effect (23). Thus,
this antagonist may have significant value in the treatment of
cancer.
In the present study, a recombinant adenovirus was
constructed, which carries a coding sequence for XAF-1 and another
sequence encoding tumor necrosis factor (TNF)-α, which induces
apoptosis similarly to XAF-1, with the 2A peptide coding sequence
(24). The anti-tumor effects of
this recombinant adenovirus was then assessed in HCC cells in
vitro. The present study provided a novel strategy for the
treatment of HCC.
Materials and methods
Tissue specimens, cell lines and
culture
A total of 56 HCC intratumor specimens and 56 paired
peritumor specimens (as controls; obtained at a distance of >10
mm from the tumor edge) were included in the present study. All
specimens were obtained from the pathological archives of Baotou
Cancer Hospital (Batou, China) and had been obtained between May
2009 and June 2014 with informed consent of the patients. The HCC
specimens had been obtained by surgical resection, immediately
frozen in liquid nitrogen and stored at −80°C prior to radiotherapy
or chemotherapy. Clinico-pathological characteristics of each
patient are listed in Table I. The
present study was approved by the Medical Ethics Committee of
Baotou Cancer Hospital (Batou, China).
 | Table IAssociation of XAF-1 mRNA with
clinico-pathological characteristics of hepatocellular carcinoma
patients [mean age, 53.4±10.3 for XAF-1 levels ≥1 and 51.5±9.6
years for XAF-1 levels <1 (P=0.7620)]. |
Table I
Association of XAF-1 mRNA with
clinico-pathological characteristics of hepatocellular carcinoma
patients [mean age, 53.4±10.3 for XAF-1 levels ≥1 and 51.5±9.6
years for XAF-1 levels <1 (P=0.7620)].
| Characteristic | XAF-1 mRNA levels
| P-value |
|---|
| ≥1 (n=20) | <1 (n=36) |
|---|
| Age (years) | | | 0.2012 |
| ≥50 | 13 | 17 | |
| <50 | 7 | 19 | |
| HBsAg | | | 0.1878 |
| Negative | 2 | 3 | |
| Positive | 8 | 13 | |
| AFP | | | 0.1385 |
| ≥200 ng/ml | 3 | 7 | |
| <200 ng/ml | 7 | 9 | |
| Tumor size | | | 0.0406 |
| ≥5 cm | 3 | 10 | |
| <5 cm | 7 | 6 | |
| BCLC stage | | | 0.0470 |
| 0-B | 8 | 8 | |
| C-D | 2 | 8 | |
| TNM stage | | | 0.0274 |
| I+II | 8 | 7 | |
| III+IV | 2 | 9 | |
| Vascular
invasion | | | 0.0731 |
| Yes | 1 | 5 | |
| No | 9 | 11 | |
The MHCC97L, HepG2 and MHCC97H human HCC cell lines
and the L02 control liver cell line were purchased from the cell
resource center of the Chinese Academy of Medical Sciences
(Beijing, China). Each cell line was cultured in Dulbecco's
modified Eagle's medium (DMEM; Ameresco, Inc., Framingham, MA, USA)
with 10% fetal bovine serum (FBS; Invitrogen; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) at 37°C in a humidified
atmosphere containing 5% CO2.
Construction of an adenovirus
co-expressing XAF-1 and TNF-α (Ad-XAF-1&TNF-α)
The open reading frame (ORF) of human XAF-1
(NM_017523) and TNF-α (NM_000594) was amplified by polymerase chain
reaction (PCR) with primers that deleted the stop codon, and was
overlapped with a sequence encoding a 2A peptide linker (24). The overlapped XAF-1 - 2A - TNF-α
nucleotide was inserted into the pShuttle-cytomegalovirus (CMV)
vector (Qbiogene, Inc., Irvine, CA, USA) to generate the
recombinant pShuttle-CMV - XAF-1 - 2A - TNF-α. The adenovirus
Ad-XAF-1&TNF-α and the Ad-control (Ad-con) virus were enveloped
via co-transfecting the pShuttle-CMV - XAF-1 - 2A - TNF-α and the
pAdeasy-1 (the viral DNA plasmid) into 293GPG retrovirus packaging
cell line (Cell Resource Center of the Chinese Academy of Medical
Sciences) using Lipofectamine™ 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.). To co-express XAF-1 and TNF-α in HCC cells,
MHCC97L cells were infected with Ad-XAF-1&TNF-α at a
multiplicity of infection (MOI) of 1 or 10 for 2 h, followed by
culture in fresh DMEM containing 2% FBS.
RNA isolation and reverse-transcription
quantitative PCR RT-qPCR
Cellular mRNA was isolated from tissues or cell
lines using TRIzol (Invitrogen; Thermo Fisher Scientific, Inc.)
according to the manufacturer's manual, following homogenization of
tissues. RT-qPCR was performed using the One Step SYBR®
Green RT-qPCR kit (Sigma-Aldrich, St. Louis, MO, USA) following the
manufacturer's instructions. The PCR reaction conditions were as
follows: Initial denaturation, 5 min at 95°C; 40 cycles of
denaturation for 20 sec at 94°C, annealing for 20 sec at 61°C and
extension for 20 sec at 72°C; and a final extension for 5 min at
72°C. The primers for XAF-1, TNF-α and β-actin were synthesized by
Invitrogen; Thermo Fisher Scientific, Inc., and were as follows:
Forward, 5′-CCCAGGGACCTCTCTCTAATC-3′ and reverse,
5′-ATGGGCTACAGGCTTGTCACT-3′ for TNF-α; forward,
5′-AGAATTCCCCATTCAGTAAG-3′ and reverse, 5′-GTGTAAGGAAGTGGTTCAGT-3′
for XAF-1; and forward, 5′-CATTAAGGAGAAGCTGTGCT-3′ and reverse,
5′-GTTGAAGGTAGTTTCGTGGA-3′ for β-actin. RT-qPCR was performed in an
ABI PRISM 7000 (Applied Biosystems; Thermo Fisher Scientific,
Inc.). Expression levels were normalized to the internal control
β-actin, expressed as the fold change compared with the control and
calculated using the ∆∆Ct method (25), subsequent to confirm the target PCR
product with melting curve analysis.
Western blot analysis
Intratumor or peritumor specimens from HCC patients
were homogenized prior to protein extraction. Lysis was then
performed with a Cell Lysis and Protein Extraction kit (Thermo
Fisher Scientific, Inc.) according to the manufacturer's
instructions, followed by addition of protease inhibitor cocktail
(Sigma-Aldrich). Proteins were quanitified using the BCA Protein
assay reagent kit (Thermo Fisher Scientific, Inc.) and 25 μg
of each sample was separated by 10% sodium dodecyl sulfate
polyacrylamide gel (Thermo Fisher Scientific, Inc.) electrophoresis
and then transferred onto a nitrocellulose membrane (EMD Millipore,
Billerica, MA, USA). Non-specific binding was blocked with 2%
bovine serum albumin (Ameresco, Inc.) overnight at 4°C, and
membranes were subsequently probed with rabbit polyclonal antibody
to XAF-1 (Abcam, Cambridge, MA, USA; cat. no. ab81353; 1:500
dilution) β-actin (Abcam; cat. no. ab8227; 1:200 dilution) or TNF-α
(Cell Signaling Technology Inc., Danvers, MA, USA; cat. no. 3727;
1:500 dilution) at 4°C overnight. The membrane was finally
incubated with goat anti-rabbit horseradish peroxidase-conjugated
secondary antibody (Promega Corp., Madison, WI, USA; cat. no.
W4011) at 4°C for 2 h, and antibodies were visualized using an
enhanced chemiluminescence detection system (GE Healthcare, Little
Chalfont, UK) following the manufacturer's instructions. The images
of the blots were captured on a UVP BioSpectrum 500 imaging system
(UVP, LLC, Upland, CA, USA) and the bands were analyzed using Image
J (imagej.nih.gov/ij/). The protein levels
of XAF-1 or TNF-α were expressed as a percentage to β-actin.
Apoptosis assay via Annexin V-fluorescein
isothiocyanate (FITC)/propidium iodide (PI) kit
The apoptosis of MHCC97L cells with or without
infection (1 MOI, 24 h) with Ad-Con, Ad-XAF-1, Ad-TNF-α or
Ad-XAF-1&TNF-α was examined with an ApoDETECT Annexin V-FITC
kit (Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol. Briefly, MHCC97L cells either without or
post-infection were incubated at 37°C for 24 h, and then were
harvested and suspended in binding buffer (5×105
cells/ml). The suspended cells were mixed with 5 μl Annexin
V-FITC and 10 μl of PI and incubated for 15 min in the dark
at room temperature. The stained cells were detected using a
FACScan flow cytometer (BD Biosciences, Franklin Lakes, NJ, USA).
The results were calculated using the CellQuest™ Pro software (BD
Biosciences) and were presented as the percentage of apoptotic
cells to total cells.
Cell proliferation assay and colony
formation assay
The proliferation of HCC cells was evaluated using a
cell counting assay and a colony formation assay. The cell counting
assay was performed as follows: Cells (103/ml) were
seeded into 12-well plates and then infected with
Ad-XAF-1&TNF-α or Ad-con virus at an MOI of 1 or 10 for 2 h,
followed by further incubation in medium for 1, 3 or 5 days. The
cells were trypsinized and the number of viable cells was counted
using a hemocytometer (Reichert, Inc., Depew, NY, USA) following
trypan blue (Thermo Fisher Scientific, Inc.) staining. For the
colony formation assay, 1,000 cells were seeded into a 12-well
plate and infected with Ad-XAF, Ad-TNF-α, Ad-XAF-1&TNF-α or
Ad-con virus at an MOI of 1 or 10 for 2 h, followed by incubation
in medium for another five days. The cell colonies were stained
with 0.5% crystal violet (Sigma-Aldrich) in methanol for 10 min and
colonies were counted on the plate by the naked eye.
Statistical analysis
Values are expressed as the mean ± standard error of
the mean. Differences between two groups were evaluated using
Student's unpaired t-test for the cell viability assay, and
the paired-samples t-test was used for comparison of
expression levels in the tumor and peritumor tissues or among the
cell lines. Statistical analysis was performed using GraphPad Prism
software (version 5; GraphPad Inc., La Jolla, CA, USA) and
P<0.05 was considered to indicate a statistically significant
difference between values.
Results
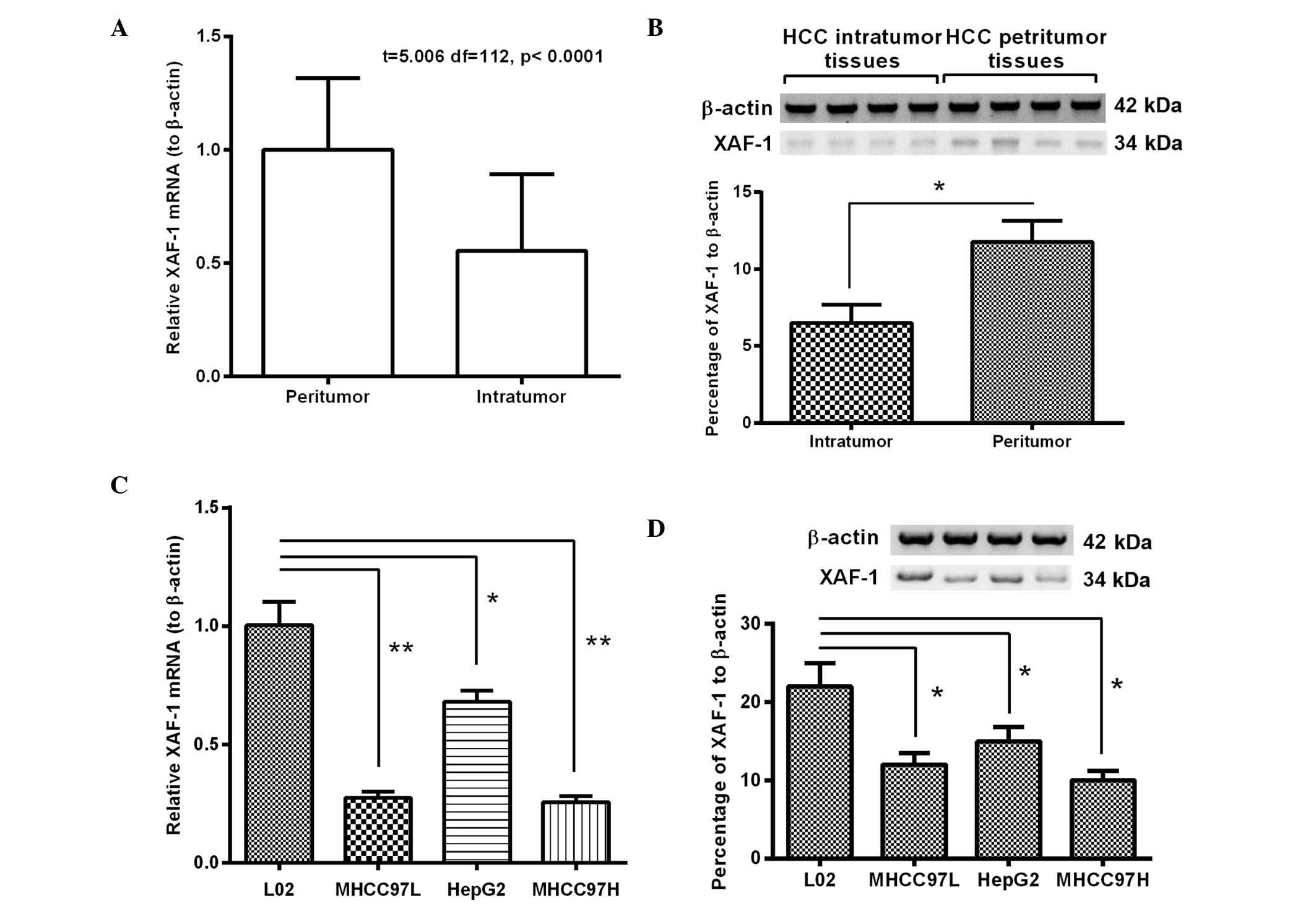
XAF-1 is downregulated in HCC specimens
and cell lines
To confirm the tumor suppressive role of XAF-1 in
HCC, its expression was determined in 56 HCC specimens and paired
peritumor tissues. As shown in Fig.
1A, compared to the levels in the peritumor tissues, the
relative XAF-1 mRNA levels (to β-actin) in the HCC specimens were
significantly reduced by ~40% (P<0.01). Furthermore, western
blot analysis confirmed a ~50% reduction of XAF-1 protein
expression in the HCC group compared with that in the peritumor
samples (P<0.001) (Fig. 1B).
Furthermore, the expression of XAF-1 in the HCC cell lines MHCC97L,
HepG2 and MHCC97H was significantly reduced at the mRNA and protein
level compared to that in the L02 human hepatic cell line
(P<0.05 or P<0.01) (Fig. 1C and
D). Thus, the significant downregulation of XAF-1 in HCC
specimens and cell lines was confirmed.
Downregulation of XAF-1 is associated
with the degree of malignancy of HCC
To assess the association of the reduced XAF-1 with
the malignant characteristics of HCC, the correlation of XAF-1
expression with clinico-pathological features, including tumor
size, Barcelona clinic liver cancer (BCLC) stage, tumor -nodes
-metastasis (TNM) stage and vascular invasion, was assessed. As
shown in Table I, there was no
significant difference in age, hepatitis B surface antigen
positivity or alpha-fetoprotein levels between the groups with
XAF-1 levels <1 and XAF-1 levels ≥1. However, XAF-1 expression
was negatively associated with the tumor size, BCLC stage, TMN
stage (P<0.05, respectively). However, the association of XAF-1
mRNA levels with vascular invasion was not significant (P>0.05).
In conclusion, these results confirmed the association of reduced
XAF-1 mRNA levels with the degree of malignancy of HCC.
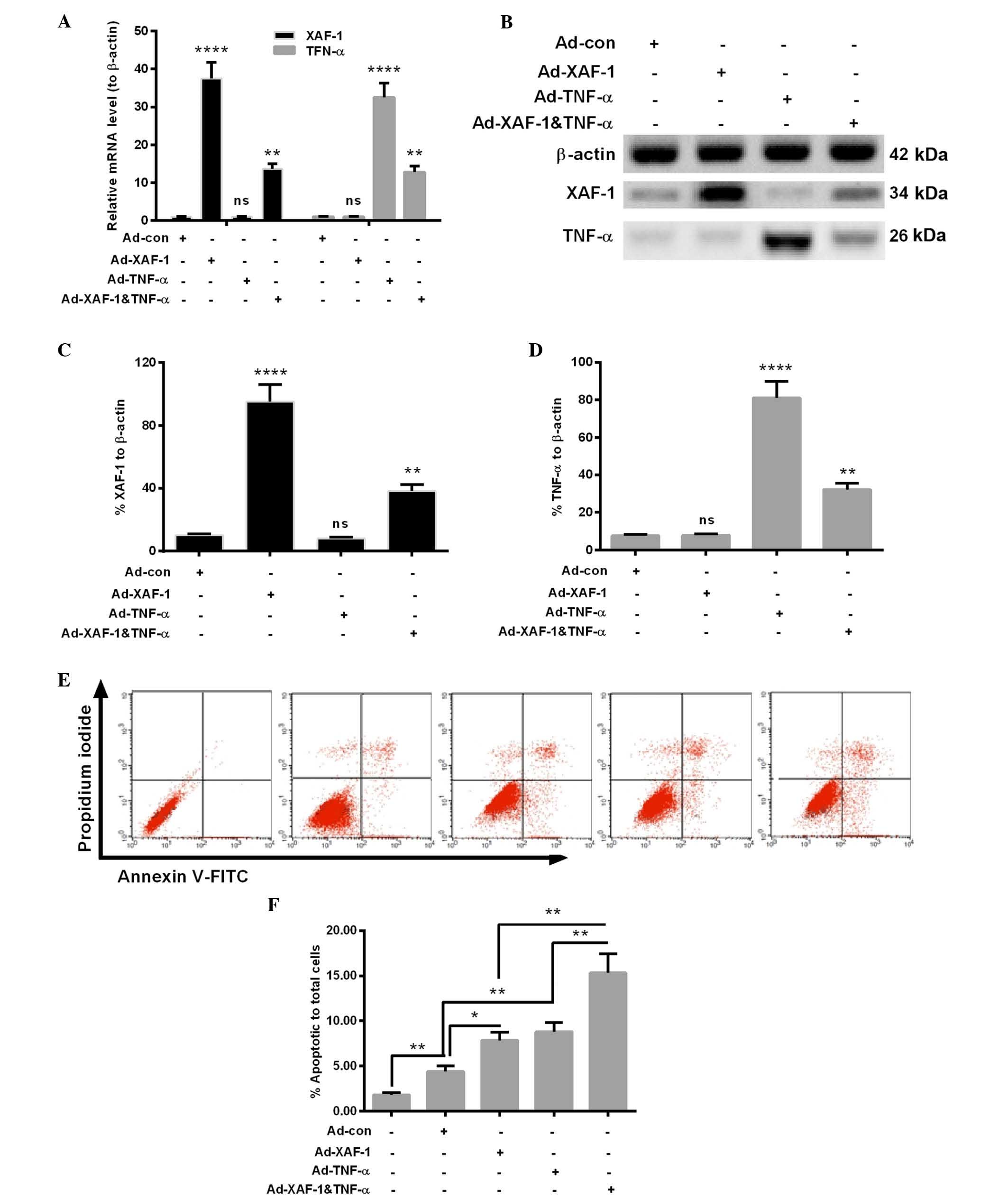
Construction of adenovirus co-expressing
XAF-1 and TNF-α
To further identify the suppressive role of XAF-1 in
HCC, an adenovirus co-expressing XAF-1 and TNF-α was constructed.
XAF-1 and TNF-α cDNA were amplified by PCR and then linked with a
2A peptide coding sequence (24).
The construction strategy of the recombinant adenovirus
Ad-XAF-1&TNF-α was illustrated in Fig. 2A. The adenovirus expressing green
fluorescence protein (Ad-con), XAF-1 (Ad-XAF-1) or TNF-α (Ad-TNF-α)
were used as controls. Each recombinant adenovirus was enveloped
via co-transfection of the respective adenoviral genomic plasmid
and the shuttle plasmid into BJ5183 bacterial cells. The efficiency
of the adenovirus to co-express XAF-1 and TNF-α was evaluated in
MHCC97L cells at an MOI of 1 or 10. At 24 h post-infection, the
mRNA levels of the two genes were significantly enhanced (P<0.01
or P<0.0001, respectively) (Fig.
2B). Furthermore, western blot analysis indicated that the
protein levels of XAF-1 and TNF-α were significantly enhanced by
the adenovirus (P<0.01, P<0.001 or P<0.0001) (Fig. 2C and D).
In addition, to compare the effects of
Ad-XAF-1&TNF-α with those of XAF-1 or TNF-α alone, MHCC97L
cells were infected with Ad-XAF-1 or Ad-TNF-α. As expected, XAF-1
was only overexpressed following infection with Ad-XAF-1, while
TNF-α was only overexpressed following infection with Ad-TNF-α at
the mRNA and protein level (P<0.001), while Ad-con had no effect
(Fig. 3). While XAF-1 as well as
TNF-α were significantly overexpressed following infection with
Ad-XAF-1&TNF-α (P<0.001), their expression levels were
significantly lower than those following infection with the
respective mono-overexpression vectors.
In addition, apoptosis in the MHCC97L cells that
were infected with 1 MOI Ad-Con, Ad-XAF-1, Ad-TNF-α or
Ad-XAF-1&TNF-α was examined. MHCC97L cells without infection
served as a blank control. As indicated in Fig. 3D and E), compared with the Ad-Con,
Ad-XAF-1 or Ad-TNF-α induced a significantly increased level of
apoptosis in MHCC97L cells (P<0.05 or P<0.01). Furthermore,
the Ad-XAF-1&TNF-α infection induced more apoptotic cells than
the infection with Ad-XAF-1 or Ad-TNF-α (P<0.01). Thus, the
co-expression of XAF-1 and TNF-α synergistically induced apoptosis
in MHCC97L cells.
Co-expression of XAF-1 and TNF-α inhibits
the growth of HCC cells
The present study then investigated the effects of
XAF-1 and TNF-α co-expression on the growth of HCC cells. The
growth of MHCC97L cells was assessed in vitro using a cell
counting assay and a colony formation assay. It was revealed that
following infection with Ad-XAF-1&TNF-α, the proliferation of
MHCC97L cells was reduced compared with that of the cells infected
with Ad-con, Ad-XAF-1 or Ad-TNF-α at either 3 days (P<0.05 or
P<0.001) or 5 days (P<0.05, P<0.01 or P<0.001)
post-infection, while infection with Ad-XAF-1 or Ad-TNF-α also
significantly inhibited the proliferation of MHCC97L cells
(P<0.05 or P<0.01) (Fig. 4A and
B). Similarly, the colony formation assay showed that the
clonogenicity of MHCC97L cells following infection with
Ad-XAF-1&TNF-α was significantly reduced compared with that
following infection with Ad-XAF-1 or Ad-TNF-α (P<0.05; Fig. 4C and D), while mono-infection still
significantly reduced the number of colonies formed (P<0.05 or
P<0.01; Fig. 4C and D). These
results indicated that co-expression of XAF-1 and TNF-α inhibited
the growth of HCC MHCC97L cells more efficiently than either
protein alone, even though their co-expression was lower than that
following infection with Ad-XAF-1 or Ad-TNF-α.
 | Figure 4Co-expression of XAF1 and TNF-α by
Ad-XAF-1&TNF-α inhibits the growth of hepatocellular carcinoma
cells. (A) Transfection with Ad-XAF-1&TNF-α significantly
reduced the number of viable MHCC97L cells compared with Ad-con,
Ad-XAF or Ad-TNF-α day five according to a Trypan blue staining
assay. (B) Statistical analysis of differences between cell numbers
in each group using the unpaired t-test. (C and D) A colony
formation assay demonstrated that Ad-XAF-1&TNF-α significantly
reduced the cologenicity of MHCC97L cells compared with Ad-con,
Ad-XAF or Ad-TNF-α. Values are expressed as the mean ± standard
error of the mean from three independent replicates.
*P<0.05, **P<0.01,
***P<0.001, or ****P<0.0001. ns, not
significant; Ad-AF-1&TNF-α, adenovirus for the co-expression of
XAF-1 and TNF-α; Ad-con, control vector; TNF, tumor necrosis
factor; XAF-1, X-linked inhibitor of apoptosis-associated factor 1;
D.P. I, days post-infection. |
Discussion
XAF-1 has been identified as a tumor suppressor gene
(8) and has been reported to be
deregulated in gastric (26),
renal (27), pancreatic (28) and esophageal (29) cancers as well as in HCCs (14). The present study reconfirmed the
downregulation of XAF-1 in HCCs at the mRNA as well as at the
protein level, which was demonstrated in HCC tissues and paired
peritumor specimens as well as in cell lines. Of note, the
downregulation of XAF-1 was associated with the degree of
malignancy of HCC, as a significant correlation of XAF-1
downregulation with the clinico-pathological characteristics of
tumor size, BCLC stage and TMN stage was identified. However, the
association of XAF-1 mRNA level with the vascular invasion was not
significant, possibly due to the small sample size.
XAF-1 has been shown to inhibit the proliferation of
lung cancer cells (30), to
suppress colon cancer growth and trigger tumor regression (31), and to induce cell apoptosis in
gastric and colorectal cancer cell lines; furthermore, XAF-1 was
reported to enhance the apoptotic effects of chemotherapeutic drugs
and TNF-related apoptosis-inducing ligand (31,32).
Recombinant adenoviral vector-mediated XAF-1 overexpression was
previously shown to significantly suppress tumor growth in gastric
and colon cancer in vitro and in vivo (14,31–34).
The present study confirmed that the co-expression of XAF-1 and
TNF-α by the Ad-XAF-1&TNF-α infection synergistically induced
apoptosis in the HCC MHCC97L cells and inhibited the proliferation
of HCC cells. To amplify the inhibitory effects of XAF-1 on HCC
cell growth, a co-expressing strategy was utilized to overexpress
XAF-1 and TNF-α by a singular adenovirus with a 2A peptide
linker.
The 2A peptide is a 'self-cleavage' peptide, which
is encoded by the foot-and-mouth disease virus. The 2A peptide
links two coding sequences in one ORF, which is transcribed into
one mRNA molecule, whereas it is translated into two different,
function-independent proteins (24). The 'self-cleavage' characteristic
of 2A peptide qualifies it to co-express two separate molecules by
same vector efficiently (35,36).
The present study was the first to constructed an adenovirus,
Ad-XAF-1&TNF-α, which co-expressed XAF-1 and TNF-α efficiently.
The expression of the two genes was significantly increased at the
mRNA as well as the protein level by infection of the
Ad-XAF-1&TNF-α into HCC MHCC97L cells. Furthermore, infection
with Ad-XAF-1&TNF-α significantly reduced the proliferation and
clonogenicity of HCC MHCC97L cells to a greater extent than
infection with the Ad-XAF-1 or Ad-TNF-α virus individually. The
present study provides a method by which XAF-1 and TNF-α were
expressed simultaneously per transcription. The co-expression
vector presents an advantage as a potential anti-tumor strategy, as
a single administration simultaneously presents two different
anti-tumor effectors in the same tumor cell.
In conclusion, the present study was the first to
construct an adenovirus which co-expressed XAF-1 and TNF-α in same
ORF and expressed them proportionally. This Ad-XAF-1&TNF-α
co-expression virus inhibited the proliferation of HCC cells more
efficiently than infection with Ad-XAF-1 or Ad-TNF-α alone,
suggesting that it may be a promising therapeutic for the treatment
of HCC.
Acknowledgments
The present study was supported by a grant from the
Baotou Bureau of Science and Technology (grant no. 2012-BT039.
References
|
1
|
Aravalli RN, Steer CJ and Cressman EN:
Molecular mechanisms of hepatocellular carcinoma. Hepatology.
48:2047–2063. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Niu J, Lin Y, Guo Z, Niu M and Su C: The
epidemiological investigation on the risk factors of hepatocellular
carcinoma: A case-control study in Southeast China. Medicine
(Baltimore). 95:e27582016. View Article : Google Scholar
|
|
3
|
Schafer DF and Sorrell MF: Hepatocellular
carcinoma. Lancet. 353:1253–1257. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Thorgeirsson SS and Grisham JW: Molecular
pathogenesis of human hepatocellular carcinoma. Nat Genet.
31:339–346. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Llovet JM, Burroughs A and Bruix J:
Hepatocellular carcinoma. Lancet. 362:1907–1917. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Huang J, Zhang X, Zhang M, Zhu JD, Zhang
YL, Lin Y, Wang KS, Qi XF, Zhang Q, Liu GZ, et al: Up-regulation of
DLK1 as an imprinted gene could contribute to human hepatocellular
carcinoma. Carcinogenesis. 28:1094–1103. 2007. View Article : Google Scholar
|
|
7
|
Huang J, Zheng DL, Qin FS, Cheng N, Chen
H, Wan BB, Wang YP, Xiao HS and Han ZG: Genetic and epigenetic
silencing of SCARA5 may contribute to human hepatocellular
carcinoma by activating FAK signaling. J Clin Invest. 120:223–241.
2010. View
Article : Google Scholar :
|
|
8
|
Iizuka N, Oka M, Yamada-Okabe H, Mori N,
Tamesa T, Okada T, Takemoto N, Tangoku A, Hamada K, Nakayama H, et
al: Comparison of gene expression profiles between hepatitis B
virus- and hepatitis C virus-infected hepatocellular carcinoma by
oligonucleotide microarray data on the basis of a supervised
learning method. Cancer Res. 62:3939–3944. 2002.PubMed/NCBI
|
|
9
|
Okada T, Iizuka N, Yamada-Okabe H, Mori N,
Tamesa T, Takemoto N, Tangoku A, Hamada K, Nakayama H, Miyamoto T,
et al: Gene expression profile linked to p53 status in hepatitis C
virus-related hepatocellular carcinoma. FEBS Lett. 555:583–590.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Oliva J, Bardag-Gorce F, French BA, Li J,
McPhaul L, Amidi F, Dedes J, Habibi A, Nguyen S and French SW:
Fat10 is an epigenetic marker for liver preneoplasia in a
drug-primed mouse model of tumorigenesis. Exp Mol Pathol.
84:102–112. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Schnabl B, Valletta D, Kirovski G and
Hellerbrand C: Zinc finger protein 267 is up-regulated in
hepatocellular carcinoma and promotes tumor cell proliferation and
migration. Exp Mol Pathol. 91:695–701. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Jiang J, Zhang H, Tang Q, Hao B and Shi R:
Expression of HIWI in human hepatocellular carcinoma. Cell Biochem
Biophys. 61:53–58. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Jain S, Singhal S, Lee P and Xu R:
Molecular genetics of hepatocellular neoplasia. Am J Transl Res.
2:105–118. 2010.PubMed/NCBI
|
|
14
|
Zhu LM, Shi DM, Dai Q, Cheng XJ, Yao WY,
Sun PH, Ding Y, Qiao MM, Wu YL, Jiang SH and Tu SP: Tumor
suppressor XAF1 induces apoptosis, inhibits angiogenesis and
inhibits tumor growth in hepatocellular carcinoma. Oncotarget.
5:5403–5415. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Henry LR, Lee HO, Lee JS, Klein-Szanto A,
Watts P, Ross EA, Chen WT and Cheng JD: Clinical implications of
fibroblast activation protein in patients with colon cancer. Clin
Cancer Res. 13:1736–1741. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Salvesen GS and Duckett CS: IAP proteins:
Blocking the road to death's door. Nat Rev Mol Cell Biol.
3:401–410. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yang YL and Li XM: The IAP family:
Endogenous caspase inhibitors with multiple biological activities.
Cell Res. 10:169–177. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ngan CY, Yamamoto H, Seshimo I, Tsujino T,
Man-i M, Ikeda JI, Konishi K, Takemasa I, Ikeda M, Sekimoto M, et
al: Quantitative evaluation of vimentin expression in tumour stroma
of colorectal cancer. Br J Cancer. 96:986–992. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhu L, Cheng X, Ding Y, Shi J, Jin H, Wang
H, Wu Y, Ye J, Lu Y, Wang TC, et al: Bone marrow-derived
myofibroblasts promote colon tumorigenesis through the
IL-6/JAK2/STAT3 pathway. Cancer Lett. 343:80–89. 2014. View Article : Google Scholar
|
|
20
|
Wu WY, Kim H, Zhang CL, Meng XL and Wu ZS:
Clinical significance of autophagic protein LC3 levels and its
correlation with XIAP expression in hepatocellular carcinoma. Med
Oncol. 31:1082014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Pan Q, Liu B, Liu J, Cai R, Liu X and Qian
C: Synergistic antitumor activity of XIAP-shRNA and TRAIL expressed
by oncolytic adenoviruses in experimental HCC. Acta Oncol.
47:135–144. 2008. View Article : Google Scholar
|
|
22
|
Plenchette S, Cheung HH, Fong WG, LaCasse
EC and Korneluk RG: The role of XAF1 in cancer. Curr Opin Investig
Drugs. 8:469–476. 2007.PubMed/NCBI
|
|
23
|
Leaman DW, Chawla-Sarkar M, Vyas K,
Reheman M, Tamai K, Toji S and Borden EC: Identification of
X-linked inhibitor of apoptosis-associated factor-1 as an
interferon-stimulated gene that augments TRAIL Apo2L-induced
apoptosis. J Biol Chem. 277:28504–28511. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Szymczak AL, Workman CJ, Wang Y, Vignali
KM, Dilioglou S, Vanin EF and Vignali DA: Correction of multi-gene
deficiency in vivo using a single 'self-cleaving' 2A peptide-based
retroviral vector. Nat Biotechnol. 22:589–594. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Schmittgen TD and Livak KJ: Analyzing
real-time PCR data by the comparative C(T) method. Nat Protoc.
3:1101–1108. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wang J, Gu Q, Li M, Zhang W, Yang M, Zou
B, Chan S, Qiao L, Jiang B, Tu S, et al: Identification of XAF1 as
a novel cell cycle regulator through modulating G(2)/M checkpoint
and interaction with checkpoint kinase 1 in gastrointestinal
cancer. Carcinogenesis. 30:1507–1516. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kempkensteffen C, Fritzsche FR, Johannsen
M, Weikert S, Hinz S, Dietel M, Riener MO, Moch H, Jung K, Krause
H, et al: Down-regulation of the pro-apoptotic XIAP associated
factor-1 (XAF1) during progression of clear-cell renal cancer. BMC
Cancer. 9:2762009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Huang J, Yao WY, Zhu Q, Tu SP, Yuan F,
Wang HF, Zhang YP and Yuan YZ: XAF1 as a prognostic biomarker and
therapeutic target in pancreatic cancer. Cancer Sci. 101:559–567.
2010. View Article : Google Scholar
|
|
29
|
Chen XY, He QY and Guo MZ: XAF1 is
frequently methylated in human esophageal cancer. World J
Gastroenterol. 18:2844–2849. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yang WT, Chen DL, Zhang FQ, Xia YC, Zhu
RY, Zhou DS and Chen YB: Experimental study on inhibition effects
of the XAF1 gene against lung cancer cell proliferation. Asian Pac
J Cancer Prev. 15:7825–7829. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Tu SP, Sun YW, Cui JT, Zou B, Lin MC, Gu
Q, Jiang SH, Kung HF, Korneluk RG and Wong BC: Tumor suppressor
XIAP-Associated factor 1 (XAF1) cooperates with tumor necrosis
factor-related apoptosis-inducing ligand to suppress colon cancer
growth and trigger tumor regression. Cancer. 116:1252–1263. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Tu SP, Liston P, Cui JT, Lin MC, Jiang XH,
Yang Y, Gu Q, Jiang SH, Lum CT, Kung HF, et al: Restoration of XAF1
expression induces apoptosis and inhibits tumor growth in gastric
cancer. Int J Cancer. 125:688–697. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Sun PH, Zhu LM, Qiao MM, Zhang YP, Jiang
SH, Wu YL and Tu SP: The XAF1 tumor suppressor induces autophagic
cell death via upregulation of Beclin-1 and inhibition of Akt
pathway. Cancer Lett. 310:170–180. 2011.PubMed/NCBI
|
|
34
|
Qi R, Gu J, Zhang Z, Yang K, Li B, Fan J,
Wang C, He Z, Qiao L, Lin Z and Liu XY: Potent antitumor efficacy
of XAF1 delivered by conditionally replicative adenovirus vector
via caspase-independent apoptosis. Cancer Gene Ther. 14:82–90.
2007. View Article : Google Scholar
|
|
35
|
De Giorgi M, Cinti A, Pelikant-Małecka I,
Chisci E, Lavitrano M, Giovannoni R and Smolenski RT: Co-expression
of functional human heme oxygenase 1, ecto-5′-nucleotidase and
ecto-nucleoside triphosphate diphosphohydrolase-1 by
'self-cleaving' 2A peptide system. Plasmid. 79:22–29. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Chng J, Wang T, Nian R, Lau A, Hoi KM, Ho
SC, Gagnon P, Bi X and Yang Y: Cleavage efficient 2A peptides for
high level monoclonal antibody expression in CHO cells. MAbs.
7:403–412. 2015. View Article : Google Scholar : PubMed/NCBI
|