Introduction
Previously, it has been reported that the aberrant
copy number variation (CNV) of the human Upf2 gene locus is
associated with neural development disorders, based on a search for
CNVs among patients with intellectual disabilities (1). Upf3b, the binding partner of Upf2, is
also associated with similar developmental disorders, and this
protein is one component of the exon junction complex (EJC) formed
on mRNA (2). The EJC, and its
associated proteins, are essential factors of the nonsense-mediated
mRNA decay (NMD) pathway, and NMD has various effects on cellular
function. Therefore, it is possible that a deficiency of them may
lead to neuro-developmental disorders.
mRNA forms messenger ribonucleoprotein particles
(mRNPs) with various factors and is exported from the nucleus to
the cytoplasm following transcription. During these processes,
various quality control systems are responsible for maintaining the
integrity of the genetic information. One of these processes is
NMD, which eliminates mRNA that contains premature termination
codons (PTCs) (3–6). PTCs may arise from a variety of
transcription errors, including nonsense mutations in the genome,
and they often cause recessive inherited disorders. Regardless of
their source, PTCs in one allele have the potential to produce
deleterious C-terminal truncated proteins, and NMD is considered to
suppress the expression of truncated proteins that possibly have
dominant negative toxicity caused by competitive inhibition of
wild-type-allele-derived proteins. During the processes of mRNA
metabolism, almost all the mRNAs that are exported from the nucleus
to the cytoplasm pass through a quality control process, so that
those containing PTCs may be eliminated to effectively maintain the
integrity of the genetic information by NMD.
The conventional NMD mechanism requires the presence
of various factors, including the SMG-1-Upf1-eRF1-eRF3 (SURF)
complex (formed with SMG1, Upf1 and eukaryotic release factors),
Upf2, Upf3b, EJC, and others. The EJC comprises RBM8A (Y14), Magoh,
eIF4A3 and other proteins, and this is formed on the mRNA via
splicing, serving as a platform for the assembly of the
NMD-associated complex (7–10) or other functional complexes. The
complex formed comprising Upf1, Upf2 and Upf3b is well established
(4,11). Upf3b and Upf2 are reported to
localize at the EJC and receive SURF when the ribosome encounters
PTCs, as determined from biochemical experiments (12).
Immunostaining experiments have revealed that EJC
and Upf3b predominantly localize in the nucleus, whereas Upf1
predominantly localizes in the cytoplasm. On the other hand,
although Upf2 has a nuclear localization signal, Upf2 is located
predominantly in the cytoplasmic perinuclear region (11,13).
The interaction of NMD factors is well established (14–16),
and NMD is considered to progress according to the following three
steps: First, Upf3b and EJC bind to mRNA molecules via RNA splicing
in the nucleus; subsequently, perinuclear Upf2 binds to Upf3b on
the exporting mRNA; and finally, following the formation of the
EJC-Upf2-mRNA complex, it interacts with the SURF complex through
Upf1 on the ribosomes arrested at a PTC, resulting in the selective
degradation of mRNAs with PTCs (4,12).
It has been reported that NMD occurs either in the cytoplasm or in
a nuclear-associated manner when newly synthesized mRNA undergoes
the first round of translation, a process known as pioneer round
translation (3,5,6,17–19).
On the other hand, the inhibitory effect of endogenous expression
of dominant-negative Upf proteins on NMD activity was assessed, and
it was proposed that the interaction between the Upf complex and
EJC occurs in the cytoplasm, rather than in a nucleus-associated
manner (20). Therefore, although
NMD occurs in the cytoplasm, the possibility of nuclear-associated
NMD during the export of mRNPs still remains.
According to previously proposed models (10,15,16),
Upf2 accumulates at the perinuclear region and forms a complex with
EJC on the exporting mRNA outside of the nuclei. However, almost
all models have been established solely on spatial observations,
and therefore have not provided any information concerning the
mechanism for Upf2 localization. If only perinuclear Upf2 proteins
are involved in forming a complex with EJC, nuclear-associated NMD
should take place close to the nucleus, perhaps immediately
following export from the nuclear pore complex. Alternatively, if
intranuclear Upf2 protein forms complexes with EJC in the nuclei,
the mRNA-EJC-Upf2 complex would appear in time for an approach by
SURF and the ribosome, even in nuclei. Indeed, at present, no
published studies have identified the location of endogenous
Upf2-EJC complex formation.
In the present study, fractionation of cell lysates
and series of immunostaining experiments were performed. Taken
together, our study using a proximity ligation in situ assay
has demonstrated that endogenous Upf2 interacts with one of the EJC
core factors, RBM8A, in the inner nucleus prior to mRNA export
through the nuclear pore, and constructs the mRNA-protein
complex.
Materials and methods
Cell culture
HeLa and A549 cells were maintained in Dulbecco's
modified Eagle's medium (Sigma-Aldrich, St. Louis, MO, USA)
supplemented with 10% fetal bovine serum and antibiotics (final
concentration, 10,000 U/ml penicillin and 10 mg/ml streptomycin;
Wako Pure Chemical Industries, Ltd., Osaka, Japan). The cells were
allowed to adhere and proliferate for ≥24 h at 37°C in 5%
CO2 prior to the following experiments.
Cellular fractionation
The preparation of nucleoplasmic and cytoplasmic
fractions was performed as previously described (21). NE-PER nuclear and cytoplasmic
extraction reagent (Pierce, Thermo Fisher Scientific, Inc.,
Rockford, IL, USA) was used according to the manufacturer's
protocol, and prepared fractions were denatured with 2X Laemmli
Sample buffer (Bio-Rad Laboratories, Inc., Hercules, CA, USA) for
western blot analysis.
Western blotting
The procedures for whole lysate preparation and
western blot analysis have been described (22). Protein concentration of the lysates
was measured by the Bradford method. In brief, denatured samples
(25 µg) were applied to 10% acrylamide gels. Following
SDS-PAGE, gels were transferred to polyvinylidene difluoride
membranes and blocked with 5% skim milk for 1 h. Then, blocked
membranes were subjected to antibody binding. The first antibodies
were rabbit anti-human Upf2 polypeptide antiserum (prepared in our
laboratory), monoclonal mouse anti-human lamin A/C antibody
(1:1,000; cat. no. sc-7292; Santa Cruz Biotechnology, Inc., Santa
Cruz, CA, USA) and monoclonal rabbit anti-human caspase 3 (1:1,000;
cat. no. 9665; Cell Signaling Technology, Inc., Danvers, MA, USA).
In addition, monoclonal mouse anti-RBM8A antibody was purchased
from Sigma-Aldrich (1:500; 4C4 clone; cat. no. Y1253). β-actin
(ACTB) was used as the reference protein, and was detected using
mouse monoclonal anti β-actin antibody purchased from Sigma-Aldrich
(1:5,000; AC 15 clone; cat. no. A5441). Incubation with primary
antibody were performed at 4°C, overnight. Primary antibodies were
detected with horseradish peroxidase-conjugated polyclonal goat
anti-mouse (1:5,000; cat. no. P0447) or anti-rabbit (1:5,000; cat.
no. P0448) secondary antibodies (Dako, Glostrup, Denmark).
Gene expression/knockdown
One day prior to the small interfering RNA (siRNA)
transfection experiments, HeLa cells were seeded on to culture
plates or dishes. The depletion of Upf2 was performed using stealth
RNA interference (RNAi) molecules (HSS178005), and the knockdown of
RBM8A was achieved using HSS115052; the two siRNAs were obtained
from Invitrogen™; Thermo Fisher Scientific, Waltham, MA, USA). Two
double-stranded molecules of the Stealth RNAi® negative
control kit (Thermo Fisher Scientific) and medium GC duplex were
used as the negative control. Transfections were performed using
Lipofectamine™ RNAiMAX transfection reagent (Thermo Fisher
Scientific), according to the manufacturer's protocol.
Immunostaining and observation of the
cells
The full details of the procedure followed for
immunostaining were previously described (23). Briefly, HeLa cells were washed
three times with phosphate-buffered saline (PBS) and fixed at room
temperature with 4% paraformaldehyde solution (Taab Laboratory
Equipment Ltd, Aldermaston, Reading, UK) in PBS for 10 min. After
washing three times with PBS, the cells were treated with 0.2%
Triton X-100 (Sigma-Aldrich)/PBS for 10 min at room temperature.
Samples were washed three times with PBS. For the ribonuclease
(RNase) treatment experiments, cells were fixed with cold ethanol
for 10 min and treated with 10 ng/ml RNaseA solution (Qiagen GmbH,
Hilden, Germany) at 37°C for 30 min, followed by three further
washes with PBS. Blocking was performed with 1% bovine serum
albumin (BSA; Wako Pure Chemical Industries, Ltd.) with gentle
agitation for 30 min at room temperature. Following this procedure,
the cells were incubated with the first antibodies, rabbit
anti-human Upf2 polypeptide antiserum prepared in our laboratory),
anti-human lamin A/C antibody (Santa Cruz Biotechnology, Inc.),
mouse anti-RBM8A antibody purchased from Sigma-Aldrich (4C4 clone)
or rabbit anti-RBM8A antibody (prepared in our laboratory) with
gentle agitation for 2 h at room temperature. Binding of the first
antibody was detected using Alexa Fluor 488- or 594-conjugated
secondary antibodies (Molecular Probes Life Technologies, Carlsbad,
CA, USA) with gentle agitation at room temperature for 60 min, and
nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI).
After washing the cells with PBS three times, ProLong®
Gold Antifade reagent (Invitrogen™; Thermo Fisher Scientific) was
used to prevent fading. Images were captured with either an
Axiovert 200 M inverted fluorescence microscope or an LSM 710
confocal point-scanning microscope (Carl Zeiss, München, Germany).
ZEN 2008 SP1.1 software (Carl Zeiss) was used to analyze signal
intensity.
Immunoprecipitation of Upf2
To confirm the association between mRNA and Upf2,
HeLa cells were collected using a cell scraper and suspended in
cold NET-2 buffer [50 mM Tris-HCl (pH 7.4), 300 mM NaCl, 0.05%
Igepal-CA630; Sigma-Aldrich]. For nuclear fractions, cells were
treated with cNE-PER nuclear and cytoplasmic extraction reagent
(Thermo Fisher Scientific, Inc.) and nuclear fractions were
collected. Subsequently, nuclear fractions were suspended in cold
NET-2 buffer. The samples were sonicated using the Vibra cell
sonicator (Sonics & Materials, Inc., Newtown, CT, USA), and the
sonicated samples were centrifuged for 15 min at 15,000 × g. The
supernatant was treated with anti-Upf2 antiserum and normal rabbit
serum as the control. Following an incubation with agitation in a
cold room, Protein G-conjugated agarose beads (Invitrogen™; Thermo
Fisher Scientific) were added to the samples, and the samples thus
obtained were again incubated in a cold room. Following sufficient
washing with NET-2 buffer, the beads were resuspended in Laemmli
sample buffer (Bio-Rad Laboratories, Inc.) with 2-mercaptoethanol
(Sigma-Aldrich). The sample was boiled, and the resultant sample
buffer solution was processed for western blotting, which was
performed as described above.
Detection of complex formation with a
proximity ligation in situ assay
The direct observation of Upf2-EJC complex formation
was performed using a Duolink® kit (Olink Bioscience,
Uppsala, Sweden; product now owned by Sigma-Aldrich) following the
manufacturer's protocol (23).
This method enables the visualization of complex formation in cells
by proximity ligation of single-stranded DNA conjugated with a
secondary antibody (24), and is
available for the complex in nuclei and cytoplasm (25). Signal can be observed when the
distance between the secondary antibodies is <40 nm. Therefore,
this method can detect not only direct protein-protein
interactions, but also complex formation, assuming that the bound
first antibodies are proximal enough. The first antibodies used in
the present study were as described in the above Immunostaining and
observation section. Images were captured with either an Axiovert
200 M inverted fluorescence microscope or an LSM 710 confocal
point-scanning microscope (Carl Zeiss, München, Germany).
Results
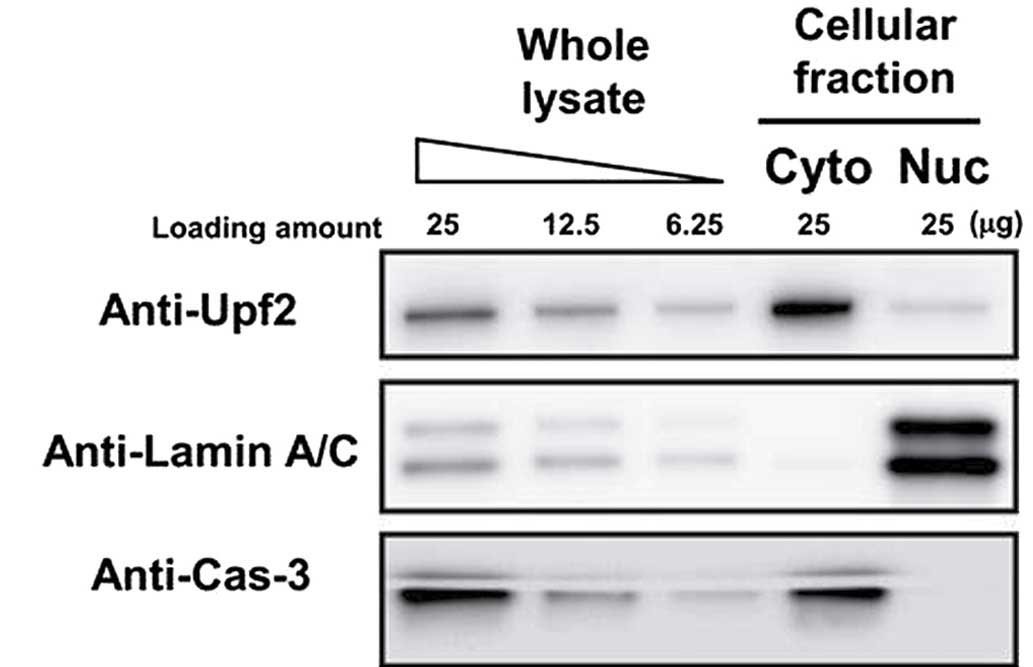
Detection of Upf2 by western blotting in
the nuclear fraction
In order to assess whether Upf2 was resident in the
nuclei, HeLa cells were fractionated into their respective nuclear
and cytoplasmic fractions. These fractions were analyzed using
western blotting. The purity of each fraction was evaluated by
blotting with anti-lamin A/C (nuclear marker) and anti-caspase 3
(cytoplasmic marker) antibodies. Almost all the lamin A/C was
detected in the nuclear fraction, and caspase 3 was predominantly
detected in the cytoplasm. Therefore, it was confirmed that the
fractionation process had been successful, and Upf2 was clearly
detected in the two fractions (Fig.
1).
Detection of Upf2 and RBM8A by
immunostaining
Subsequently, to confirm the results of western
blotting, immunostaining experiments were performed. Previous
reports have revealed the cytoplasmic and perinuclear localization
of endogenous or exogenous Upf2 (11,13).
In accordance with these reports, the results in the present study
also demonstrated that Upf2 is predominantly localized to the
cytoplasmic perinuclear region, although a limited quantity of
signal was observed in the nucleus (Fig. 2A). Subsequently, knockdown
experiments were performed to confirm the specificity of the
obtained signals. The Upf2-specific siRNA, prepared in our own
laboratory, successfully depleted the Upf2 signals in the
perinuclear and intranuclear regions (Fig. 2A). The knockdown efficiency was
confirmed by western blotting of the siRNA-treated and control
siRNA-treated cells (Fig. 2B).
These findings suggested that the signals from immunostaining
observed in the present study had originated from the Upf2
molecules. Thus, it was deduced that Upf2 proteins exist not only
in the perinuclear region, but also in the nucleoplasmic
region.
Upf2 binds to RNA complex
To analyze which factors bind to the RNA molecule,
RNase treatments are available and have been used in a number of
previous studies (26–28). To examine whether Upf2 protein
binds to RNA molecules in cells, methanol-fixed cells were treated
with RNaseA solution. Following washing of the cells, cells were
stained with anti-lamin A/C and anti-Upf2 antibodies, as described
in the Materials and methods section. In addition, RBM8A was also
stained as a control for the RNase treatment. As shown in Fig. 3, the majority of Upf2 in the
cytoplasm and the nucleoplasm disappeared on RNase treatment. This
suggested that appreciable quantities of Upf2 bind to the mRNA
molecules in the nucleoplasm, as found for the cytoplasm and the
perinuclear region. The signal from lamin A/C remained, and this
clearly demonstrated that the structural integrity of the nucleus
was not affected by the RNase treatment.
The presence of Upf2-binding RBM8A in the nuclear
fraction was subsequently investigated. Following fractionation,
Upf2 protein was immunopurified using a specific antibody. RBM8A
was detected to a limited extent in the immunoprecipitate of the
nuclear fraction, similarly to the cytoplasmic fraction (Fig. 4).
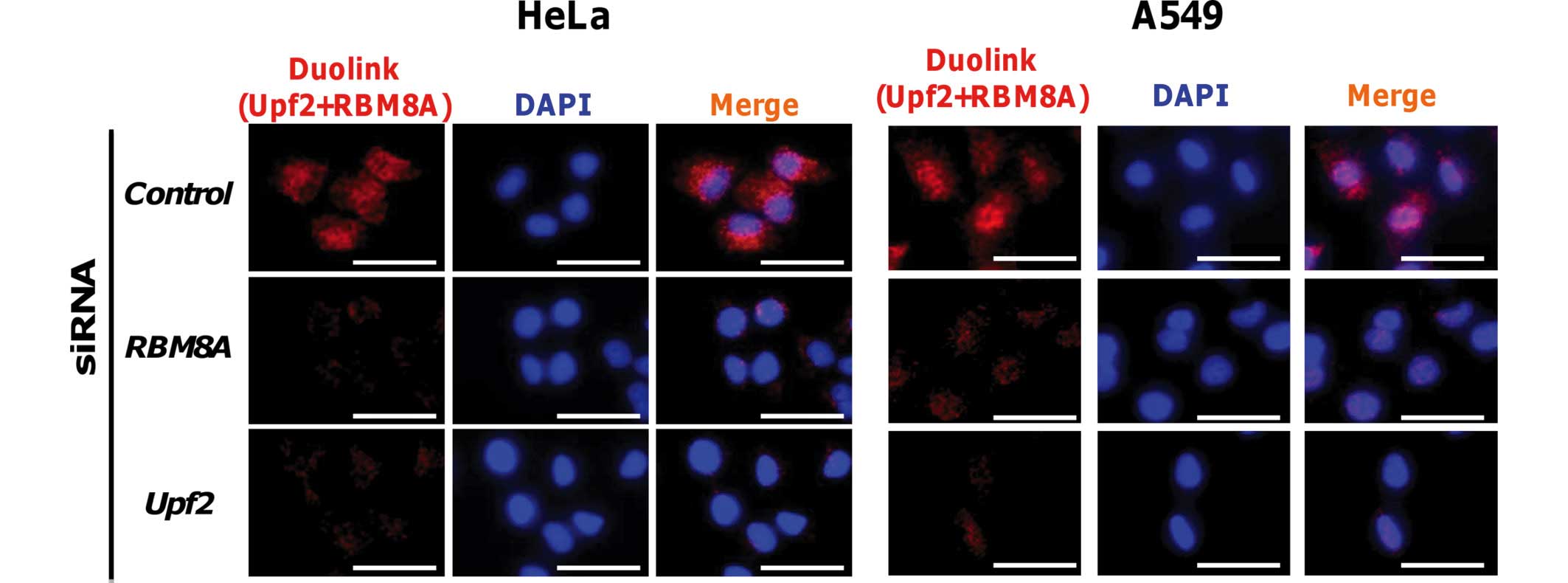
Detection of complex formation between
Upf2 and RBM8A using a proximity ligation in situ assay
Finally, to verify the interaction between Upf2 and
EJC in the nucleoplasm, a proximity ligation in situ assay
with rabbit anti-Upf2 and mouse anti-RBM8A antibodies (a component
of EJC) was performed (7–10,29).
Signals from the proximity ligation in situ assay were
detected under a fluorescence microscope from the nuclei and the
cytoplasm. In addition, knockdown of either the Upf2 or the RBM8A
gene resulted in a reduction in signal intensity (Fig. 5). Under a confocal laser scanning
microscope, sliced images were obtained, and the images revealed
the nuclear-localized signals in addition to cytoplasmic signals
(data not shown). These findings were not cell-type-specific, since
similar results were obtained with human A549 cells under identical
conditions. These results suggested that the Upf2 protein resides
proximally to RBM8A in the nuclei and cytoplasm, and is included in
the EJC.
Discussion
Previous reports have demonstrated that Upf2 binding
at the perinuclear region efficiently promotes NMD before
translation. Although Upf2 has a putative nuclear localization
signal (NLS) sequence and is localized to the perinuclear region,
whether Upf2 is present in the nucleus remains unclear. Thus, the
current study investigated the presence of Upf2 in the nucleus. The
data suggested that nuclear Upf2 co-localizes with mRNPs in the
nucleus. Thus. the previously proposed model (10,15,16),
which included cytoplasmic binding, requires the addition of a
nucleoplasmic fraction.
Taken together, our results suggest that nuclear
Upf2 co-localizes with mRNPs in the nucleus. The previously
proposed model (10,15,16),
which included cytoplasmic binding, therefore requires the addition
of a nucleoplasmic fraction. Since Upf2-associated NMD occurs in
the cytoplasm (20), nuclear
complex formation may not be associated with the cytoplasmic NMD
reaction. In addition, the distribution of the Duolink signal did
not perfectly correlate with the localization of Upf2, and complex
formation and cytoplasmic Upf2 are able to exist without complex
formation with EJC. Therefore, the mechanism that would account for
the binding of Upf2 to the EJC in the nucleus has yet to be
elucidated. Essentially, the molecular function of Upf2 has not
been firmly established, other than the requirement for NMD
activity. Since NMD occurs in the cytoplasm (20), given its nuclear function,
additional functions for Upf2 may be assumed. In a transcriptome
analysis, depletion of Upf2 was demonstrated to cause physiological
changes in various genes without PTC (30). Therefore, Upf2 may have additional
functions that would be required for the proper development of the
human neural system. Additionally, the quantity of Upf2 that
resides in the nuclei has yet to be fully established, since there
is a possibility that the western blots and immunoprecipitation
data also contained nuclear-flanked Upf2. Therefore, further
investigations are revealed to reveal additional functions of Upf2,
including its role in the nucleus.
Acknowledgments
This work was supported by grants from the Kanazawa
Medical University (nos. C2014-3, S2014-15), a grant from the
Strategic Research Project (no. H2012-16 [S1201022]), from Kanazawa
Medical University, Ministry of Education, Culture, Sports, Science
and Technology, Japan (KAKENHI no. #25460376) and from the Sumitomo
Science Foundation in Japan.
References
|
1
|
Nguyen LS, Kim HG, Rosenfeld JA, Shen Y,
Gusella JF, Lacassie Y, Layman LC, Shaffer LG and Gécz J:
Contribution of copy number variants involving nonsense-medated
mRNA decay pathway genes to neuro-developmental disorders. Hum Mol
Genet. 22:1816–1825. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Tarpey PS, Raymond FL, Nguyen LS,
Rodriguez J, Hackett A, Vandeleur L, Smith R, Shoubridge C, Edkins
S, Stevens C, et al: Mutations in UPF3B, a member of the
nonsense-mediated mRNA decay complex, cause syndromic and
nonsyndromic mental retardation. Nat Genet. 39:1127–1133. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bühler M, Wilkinson MF and Mühlemann O:
Intranuclear degradation of nonsense codon-containing mRNA. EMBO
Rep. 3:646–651. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chamieh H, Ballut L, Bonneau F and Le Hir
H: NMD factors UPF2 and UPF3 bridge UPF1 to the exon junction
complex and stimulate its RNA helicase activity. Nat Struct Mol
Biol. 15:85–93. 2008. View
Article : Google Scholar
|
|
5
|
Mühlemann O, Eberle AB, Stalder L and
Zamudio Orozco R: Recognition and elimination of nonsense mRNA.
Biochim Biophys Acta. 1779:538–549. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Maquat LE, Tarn WY and Isken O: The
pioneer round of translation: Features and functions. Cell.
142:368–374. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gehring NH, Neu-Yilik G, Schell T, Hentze
MW and Kulozik AE: Y14 and hUpf3b form an NMD-activating complex.
Mol Cell. 11:939–949. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gehring NH, Lamprinaki S, Hentze MW and
Kulozik AE: The hierarchy of exon-junction complex assembly by the
spliceosome explains key features of mammalian nonsense-mediated
mRNA decay. PLoS Biol. 7:e10001202009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kataoka N, Yong J, Kim VN, Velazquez F,
Perkinson RA, Wang F and Dreyfuss G: Pre-mRNA splicing imprints
mRNA in the nucleus with a novel RNA-binding protein that persists
in the cytoplasm. Mol Cell. 6:673–682. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kim VN, Kataoka N and Dreyfuss G: Role of
the nonsense-mediated decay factor hUpf3 in the splicing-dependent
exon-exon junction complex. Science. 293:1832–1836. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Serin G, Gersappe A, Black JD, Aronoff R
and Maquat LE: Identification and characterization of human
orthologues to Saccharomyces cerevisiae Upf2 protein and Upf3
protein (Caenorhabditis elegans SMG-4). Mol Cell Biol. 21:209–223.
2001. View Article : Google Scholar
|
|
12
|
Kashima I, Yamashita A, Izumi N, Kataoka
N, Morishita R, Hoshino S, Ohno M, Dreyfuss G and Ohno S: Binding
of a novel SMG-1-Upf1-eRF1-eRF3 complex (SURF) to the exon junction
complex triggers Upf1 phosphorylation and nonsense-mediated mRNA
decay. Genes Dev. 20:355–367. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lykke-Andersen J, Shu MD and Steitz JA:
Human Upf proteins target an mRNA for nonsense-mediated decay when
bound downstream of a termination codon. Cell. 103:1121–1131. 2000.
View Article : Google Scholar
|
|
14
|
Kim VN, Yong J, Kataoka N, Abel L, Diem MD
and Dreyfuss G: The Y14 protein communicates to the cytoplasm the
position of exon-exon junctions. EMBO J. 20:2062–2068. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Le Hir H, Gatfield D, Izaurralde E and
Moore MJ: The exon-exon junction complex provides a binding
platform for factors involved in mRNA export and nonsense-mediated
mRNA decay. EMBO J. 20:4987–4997. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Schell T, Kulozik AE and Hentze MW:
Integration of splicing, transport and translation to achieve mRNA
quality control by the nonsense-mediated decay pathway. Genome
Biol. 3:REVIEWS10062002. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ishigaki Y, Li X, Serin G and Maquat LE:
Evidence for a pioneer round of mRNA translation: mRNAs subject to
nonsense-mediated decay in mammalian cells are bound by CBP80 and
CBP20. Cell. 106:607–617. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chang YF, Imam JS and Wilkinson MF: The
nonsense-mediated decay RNA surveillance pathway. Annu Rev Biochem.
76:51–74. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Maquat LE: Nonsense-mediated mRNA decay:
Splicing, translation and mRNP dynamics. Nat Rev Mol Cell Biol.
5:89–99. 2004. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Singh G, Jakob S, Kleedehn MG and
Lykke-Andersen J: Communication with the exon-junction complex and
activation of nonsense-mediated decay by human Upf proteins occur
in the cytoplasm. Mol Cell. 27:780–792. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhao X, Nogawa A, Matsunaga T, Takegami T,
Nakagawa H and Ishigaki Y: Proteasome inhibitors and knockdown of
SMG1 cause accumulation of Upf1 and Upf2 in human cells. Int J
Oncol. 44:222–228. 2014.
|
|
22
|
Ishigaki Y, Nakamura Y, Tatsuno T,
Hashimoto M, Shimasaki T, Iwabuchi K and Tomosugi N: Depletion of
RNA-binding protein RBM8A (Y14) causes cell cycle deficiency and
apoptosis in human cells. Exp Biol Med (Maywood). 238:889–897.
2013. View Article : Google Scholar
|
|
23
|
Ishigaki Y, Nakamura Y, Tatsuno T,
Hashimoto M, Iwabuchi K and Tomosugi N: RNA binding protein RBM8A
(Y14) and MAGOH localize to centrosome in human A549 cells.
Histochem Cell Biol. 141:101–109. 2014. View Article : Google Scholar
|
|
24
|
Söderberg O, Gullberg M, Jarvius M,
Ridderstråle K, Leuchowius KJ, Jarvius J, Wester K, Hydbring P,
Bahram F, Larsson LG and Landegren U: Direct observation of
individual endogenous protein complexes in situ by proximity
ligation. Nat Methods. 3:995–1000. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hervouet E, Lalier L, Debien E, Cheray M,
Geairon A, Rogniaux H, Loussouarn D, Martin SA, Vallette FM and
Cartron PF: Disruption of Dnmt1/PCNA/UHRF1 interactions promotes
tumorigenesis from human and mice glial cells. PLoS One.
5:e113332010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Spector DL, Fu XD and Maniatis T:
Associations between distinct pre-mRNA splicing components and the
cell nucleus. EMBO J. 10:3467–3481. 1991.PubMed/NCBI
|
|
27
|
den Engelsman J, Bennink EJ, Doerwald L,
Onnekink C, Wunderink L, Andley UP, Kato K, de Jong WW and Boelens
WC: Mimicking phosphorylation of the small heat-shock protein
alphaB-crystallin recruits the F-box protein FBX4 to nuclear SC35
speckles. Eur J Biochem. 271:4195–4203. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zirwes RF, Eilbracht J, Kneissel S and
Schmidt-Zachmann MS: A novel helicase-type protein in the
nucleplus: Protein NOH61. Mol Cell Biol. 11:1153–1167. 2000.
View Article : Google Scholar
|
|
29
|
Schmidt U, Richter K, Berger AB and
Lichter P: In vivo BiFC analysis of Y14 and NXF1 mRNA export
complexes: Preferential localization within and around SC35
domains. J Cell Biol. 172:373–381. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wittmann J, Hol EM and Jäck HM: hUPF2
silencing identifies physiologic substrates of mammalian
nonsense-mediated mRNA decay. Mol Cell Biol. 26:1272–1287. 2006.
View Article : Google Scholar : PubMed/NCBI
|