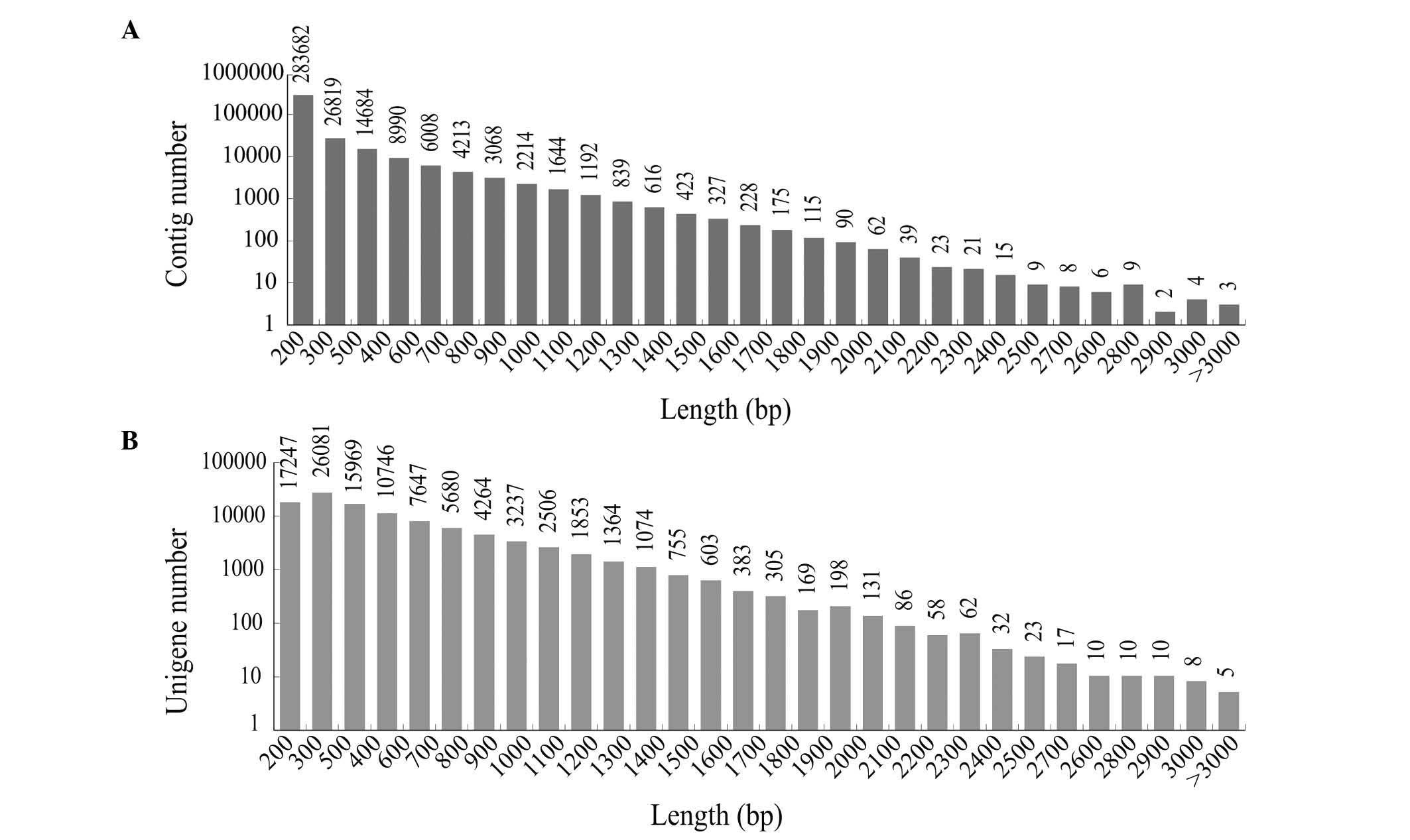
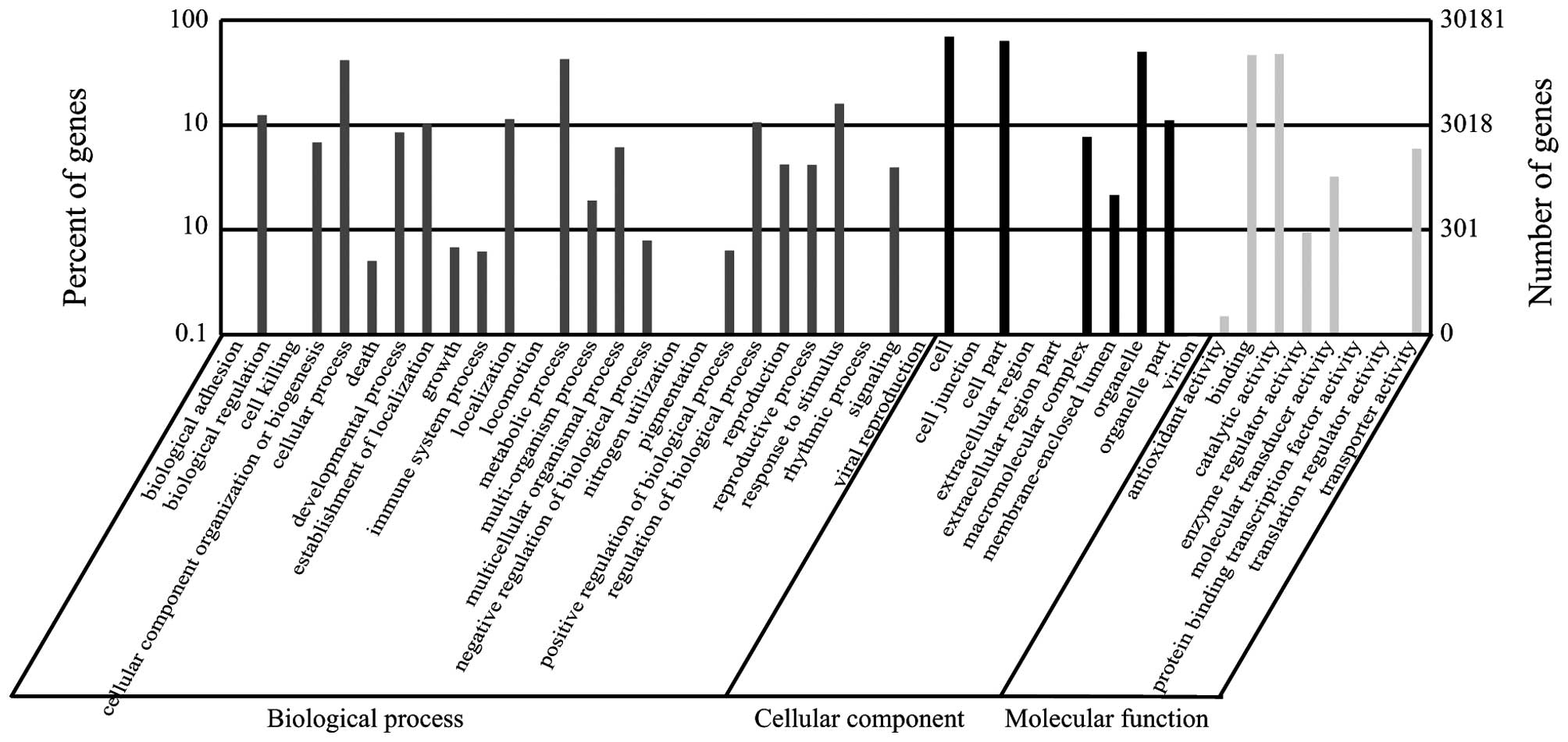
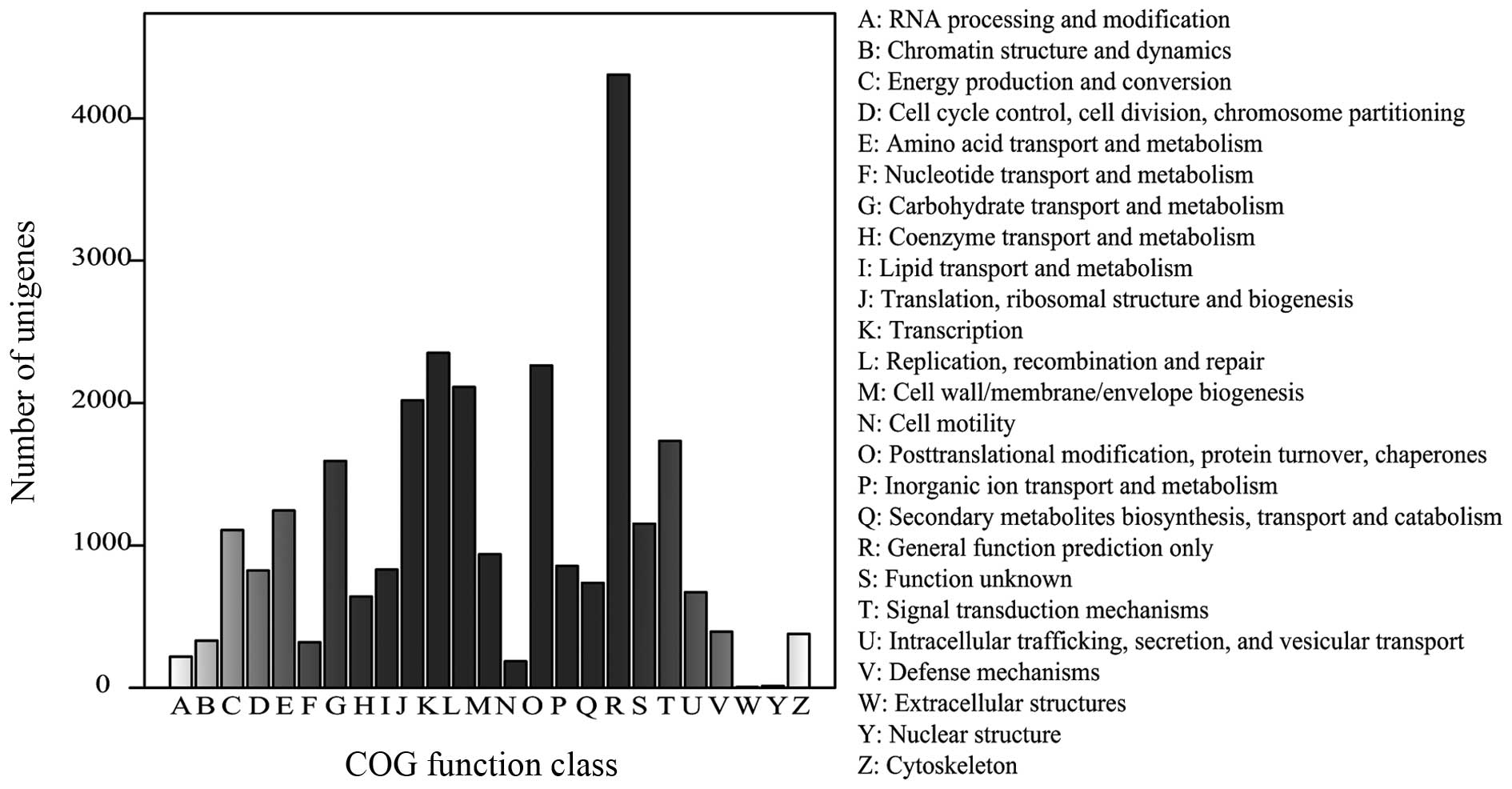
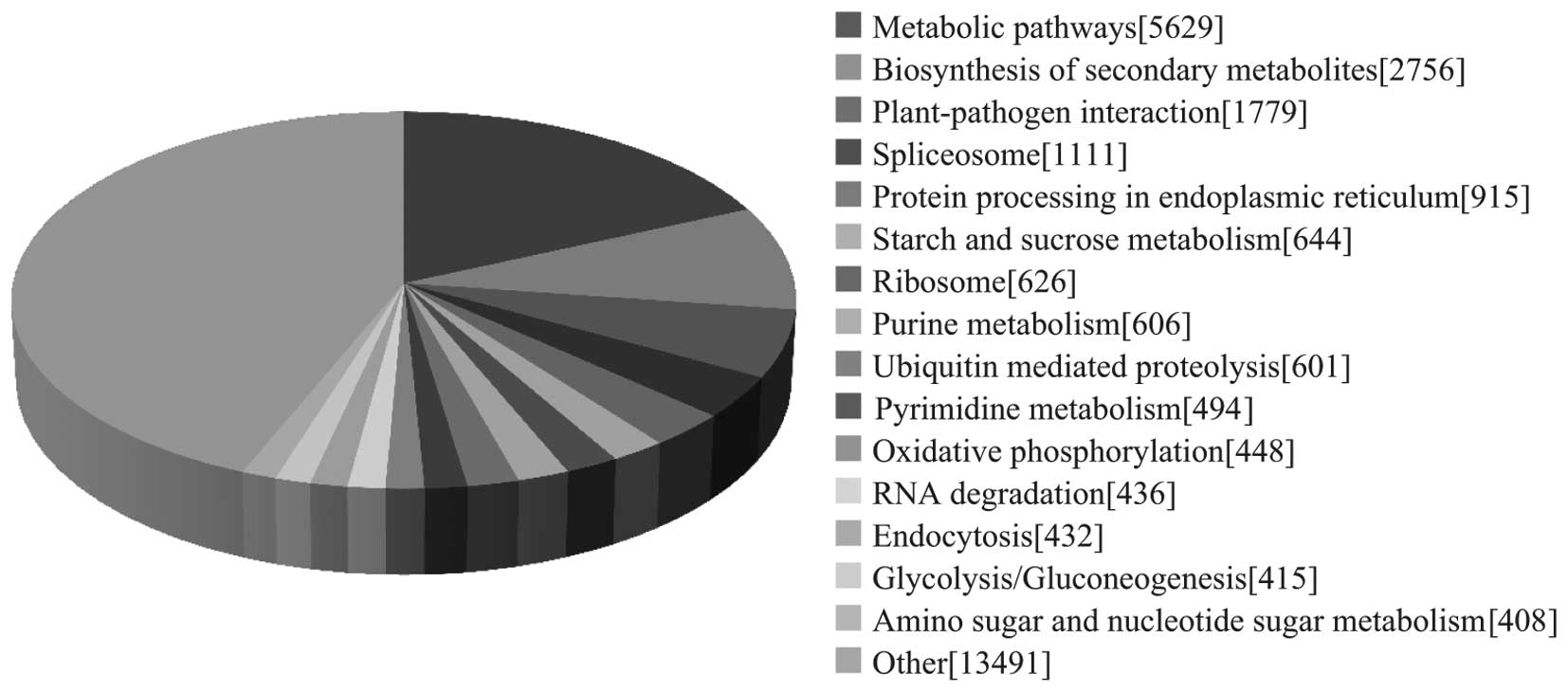
|
1
|
Briskin DP: Medicinal plants and
phytomedicines. Linking plant biochemistry and physiology to human
health. Plant Physiol. 124:507–514. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kim YJ, Zhang D and Yang DC: Biosynthesis
and biotechnological production of ginsenosides. Biotechnol Adv.
33:717–735. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Smith I, Williamson EM, Putnam S,
Farrimond J and Whalley BJ: Effects and mechanisms of ginseng and
ginsenosides on cognition. Nutr Rev. 72:319–333. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wu D, Austin RS, Zhou S and Brown D: The
root transcriptome for North American ginseng assembled and
profiled across seasonal development. BMC Genomics. 14:5642013.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Munns R, Passioura JB, Guo J, Chazen O and
Cramer GR: Water relations and leaf expansion: Importance of time
scale. J Exp Bot. 51:1495–1504. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wei YC, Chen CB, Li YB, et al: Ginseng
development practical manual. Jilin Science and Technology Bureau;
Changchun: 2010
|
|
7
|
Peng Y, Lin W, Cai W and Arora R:
Overexpression of a Panax ginseng tonoplast aquaporin alters salt
tolerance, drought tolerance and cold acclimation ability in
transgenic Arabidopsis plants. Planta. 226:729–740. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Pulla RK, Kim YJ, Parvin S, Shim JS, Lee
JH, Kim YJ, In JG, Senthil KS and Yang DC: Isolation of
S-adenosyl-L-methionine synthetase gene from Panax ginseng C.A.
meyer and analysis of its response to abiotic stresses. Physiol Mol
Biol Plants. 15:267–275. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lee SK: Fusarium species associated with
ginseng (Panax ginseng) and their role in the root-rot of ginseng
plants. Res Plant Dis. 10:248–259. 2004. View Article : Google Scholar
|
|
10
|
Li Y, Liu SL, Huang XF and Ding WL:
Allelopathy of ginseng root exudates on pathogens of ginseng. Acta
Ecological Sinica. 29:161–167. 2009.
|
|
11
|
Lee OR, Pulla RK, Kim YJ, Balusamy SR and
Yang DC: Expression and stress tolerance of PR10 genes from Panax
ginseng C. A. Meyer. Mol Biol Rep. 39:2365–7234. 2012. View Article : Google Scholar
|
|
12
|
Gupta P, Goel R, Pathak S, Srivastava A,
Singh SP, Sangwan RS, Asif MH and Trivedi PK: De novo assembly,
functional annotation and comparative analysis of Withania
somnifera leaf and root transcriptomes to identify putative genes
involved in the withanolides biosynthesis. PLoS One. 8:e627142013.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Margulies M, Egholm M, Altman WE, Attiya
S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z, et
al: Genome sequencing in microfabricated high-density picolitre
reactors. Nature. 437:376–380. 2005.PubMed/NCBI
|
|
14
|
Wang Z, Gerstein M and Snyder M: RNA-Seq:
A revolutionary tool for transcriptomics. Nat Rev Genet. 10:57–63.
2009. View
Article : Google Scholar
|
|
15
|
Chen S, Luo H, Li Y, Sun Y, Wu Q, Niu Y,
Song J, Lv A, Zhu Y, Sun C, et al: 454 EST analysis detects genes
putatively involved in ginsenoside biosynthesis in Panax ginseng.
Plant Cell Rep. 30:1593–1601. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Choi DW, Jung J, Ha YI, Park HW, In DS,
Chung HJ and Liu JR: Analysis of transcripts in methyl
jasmonate-treated ginseng hairy roots to identify genes involved in
the biosynthesis of ginsenosides and other secondary metabolites.
Plant Cell Rep. 23:557–566. 2005. View Article : Google Scholar
|
|
17
|
Fleige S and Pfaffl MW: RNA integrity and
the effect on the real-time qRT-PCR performance. Mol Aspects Med.
27:126–139. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Metzker ML: Sequencing technologies-the
next generation. Nat Rev Genet. 11:31–46. 2010. View Article : Google Scholar
|
|
19
|
King M, Reeve W, Van der Hoek MB, Williams
N, McComb J, O'Brien PA and Hardy GE: Defining the
phosphite-regulated transcriptome of the plant pathogen
Phytophthora cinnamomi. Mol Genet Genomics. 284:425–435. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Jiang Q, Wang F, Tan HW, Li MY, Xu ZS, Tan
GF and Xiong AS: De novo transcriptome assembly, gene annotation,
marker development and miRNA potential target genes validation
under abiotic stresses in Oenanthe javanica. Mol Genet Genomics.
290:671–683. 2015. View Article : Google Scholar
|
|
21
|
Yu M, Yu J, Gu C, Nie Y, Chen Z, Yin X and
Liu Y: De novo sequencing and transcriptome analysis of
Ustilaginoidea virens by using Illumina paired-end sequencing and
development of simple sequence repeat markers. Gene. 547:202–210.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Tsanakas GF, Manioudaki ME, Economou AS
and Kalaitzis P: De novo transcriptome analysis of petal senescence
in Gardenia jasminoides Ellis. BMC Genomics. 15:5542014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Grabherr MG, Haas BJ, Yassour M, Levin JZ,
Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, et
al: Full-length transcriptome assembly from RNA-Seq data without a
reference genome. Nat Biotechnol. 15:644–652. 2011. View Article : Google Scholar
|
|
24
|
Wang Z, Zhang J, Jia C, Liu J, Li Y, Yin
X, Xu B and Jin Z: De novo characterization of the banana root
transcriptome and analysis of gene expression under Fusarium
oxysporum f. sp Cubense tropical race 4 infection. BMC Genomics.
13:6502012. View Article : Google Scholar
|
|
25
|
Lee Y, Tsai J, Sunkara S, Karamycheva S,
Pertea G, Sultana R, Antonescu V, Chan A, Cheung F and Quackenbush
J: The TIGR Gene Indices: Clustering and assembling EST and known
genes and integration with eukaryotic genomes. Nucleic Acids Res.
33(Database Issue): D71–D74. 2005. View Article : Google Scholar :
|
|
26
|
Mortazavi A, Williams BA, McCue K,
Schaeffer L and Wold B: Mapping and quantifying mammalian
transcriptomes by RNA-Seq. Nat Methods. 5:621–628. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Conesa A, Götz S, García-Gómez JM, Terol
J, Talón M and Robles M: Blast2GO: A universal tool for annotation,
visualization and analysis in functional genomics research.
Bioinformatics. 15:3674–3676. 2005. View Article : Google Scholar
|
|
28
|
Harris MA, Clark J, Ireland A, Lomax J,
Ashburner M, Foulger R, Eilbeck K, Lewis S, Marshall B, Mungall C,
et al: The gene ontology (GO) database and informatics resource.
Nucleic Acids Res. 32(Database Issue): D258–D261. 2004. View Article : Google Scholar
|
|
29
|
Kanehisa M, Goto S, Kawashima S, Okuno Y
and Hattori M: The KEGG resource for deciphering the genome.
Nucleic Acids Res. 32(Database Issue): D277–D280. 2004. View Article : Google Scholar :
|
|
30
|
Tatusov RL, Natale DA, Garkavtsev IV,
Tatusova TA, Shankavaram UT, Rao BS, Kiryutin B, Galperin MY,
Fedorova ND and Koonin EV: The COG database: New developments in
phylogenetic classification of proteins from complete genomes.
Nucleic Acids Res. 29:22–28. 2001. View Article : Google Scholar :
|
|
31
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta DeltaC (T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
32
|
Hudson ME: Sequencing breakthroughs for
genomic ecology and evolutionary biology. Mol Ecol Resour. 8:3–17.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wagner GP, Kin K and Lynch VJ: Measurement
of mRNA abundance using RNA-seq data: RPKM measure is inconsistent
among samples. Theory Biosci. 131:281–285. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kim SI, Kweon SM, Kim EA, Kim JY, Kim S,
Yoo JS and Park YM: Characterization of RNase-like major storage
protein from the ginseng root by proteomic approach. J Plant
Physiol. 161:837–845. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Sels J, Mathys J, De Coninck BM, Cammue BP
and De Bolle MF: Plant pathogenesis-related (PR) proteins: A focus
on PR peptides. Plant Physiol Biochem. 46:941–950. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Van Loon LC, Rep M and Pieterse CM:
Significance of inducible defense-related proteins in infected
plants. Annu Rev Phytopathol. 44:135–162. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Sticher L, Mauch-Mani B and Métraux JP:
Systemic acquired resistance. Annu Rev Phytopathol. 35:235–270.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
van Loon LC: Pathogenesis-related
proteins. Plant Mol Biol. 4:111–116. 1985. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Somssich IE, Schmelzer E, Kawalleck P and
Hahlbrock K: Gene structure and in situ transcript localization of
pathogenesis-related protein 1 in parsley. Mol Gen Genet.
213:93–98. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Moiseyev GP, Fedoreyeva LI, Zhuravlev YN,
Yasnetskaya E, Jekel PA and Beintema JJ: Primary structures of two
ribo-nucleases from ginseng calluses. New members of the PR-10
family of intracellular pathogenesis-related plant proteins. FEBS
Lett. 28:207–210. 1997. View Article : Google Scholar
|
|
41
|
Purev M, Kim YJ, Kim MK, Pulla RK and Yang
DC: Isolation of a novel catalase (Cat1) gene from Panax ginseng
and analysis of the response of this gene to various stresses.
Plant Physiol Biochem. 48:451–460. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Sathiyaraj G, Lee OR, Parvin S,
Khorolragchaa A, Kim YJ and Yang DC: Transcript profiling of
antioxidant genes during biotic and abiotic stresses in Panax
ginseng C. A. Meyer. Mol Biol Rep. 38:2761–2769. 2011. View Article : Google Scholar
|
|
43
|
Hernandez JA, Jimenez A, Mullineaux P and
Sevilia F: Tolerance of pea (Pisumsativum L) to long-term salt
stress is associated with induction of antioxidant defenses. Plant
Cell Environ. 23:853–862. 2000. View Article : Google Scholar
|
|
44
|
Apel K and Hirt H: Reactive oxygen
species: Metabolism, oxidative stress, and signal transduction.
Annu Rev Plant Biol. 55:373–399. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Brkljacić JM, Samardzić JT, Timotijević GS
and Maksimović VR: Expression analysis of buckwheat (Fagopyrum
esculentum Moench) metallothionein-like gene (MT3) under different
stress and physiological conditions. J Plant Physiol. 161:741–746.
2004. View Article : Google Scholar
|
|
46
|
Mohamed H and Al-Whaibi: Plant heat-shock
proteins: A mini review. Journal of King Saud University Science.
23:139–150. 2011. View Article : Google Scholar
|
|
47
|
Jung JD, Park HW, Hahn Y, Hur CG, In DS,
Chung HJ, Liu JR and Choi DW: Discovery of genes for ginsenoside
biosynthesis by analysis of ginseng expressed sequence tags. Plant
Cell Rep. 22:224–230. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Ingram J and Bartels D: The molecular
basis of dehydration tolerance in plants. Annu Rev Plant Physiol
Plant Mol Biol. 47:377–403. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Pockley AG and Muthana M: Heat shock
proteins and allograft rejection. Contrib Nephrol. 148:122–134.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Xu D, Duan X, Wang B, Hong B, Ho T and Wu
R: Expression of a late embryogenesis abundant protein gene, HVA1,
from barley confers tolerance to water deficit and salt stress in
transgenic rice. Plant physiol. 110:249–257. 1996.PubMed/NCBI
|
|
51
|
Shen L, Xu J, Dong LL, Li XW and Chen SL:
Cropping system and research strategies in Panax ginseng. Zhongguo
Zhong Yao Za Zhi. 40:3367–3373. 2015.In Chinese.
|
|
52
|
Liu Y, Zhao D, Liu M, Hu CY, Li Y and Ding
WL: Investigation of pests and diseases occurrence and pesticides
application in main producing areas of Panax ginseng. Chinese
Agricultural Science Bulletin. 30:294–298. 2014.
|