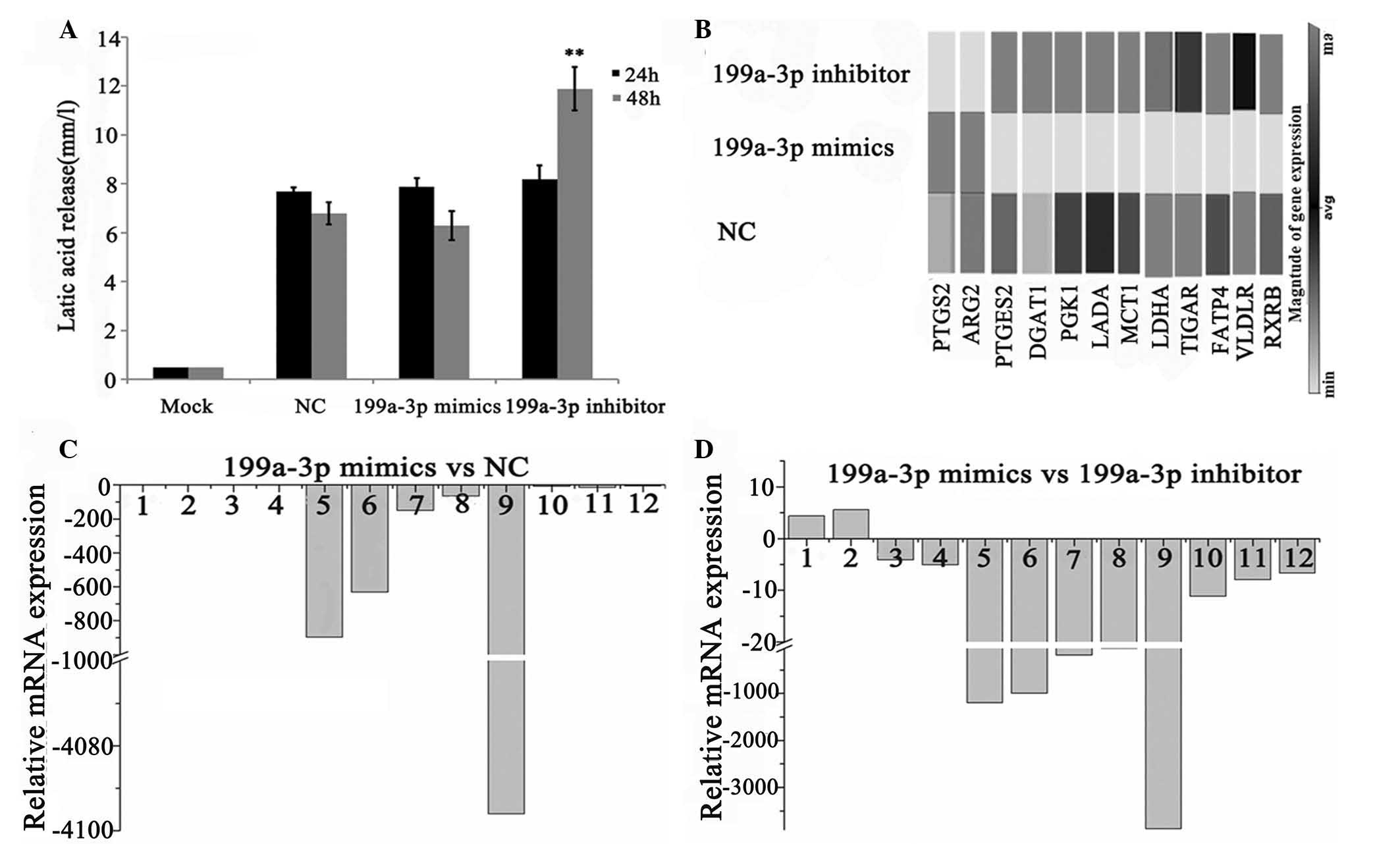
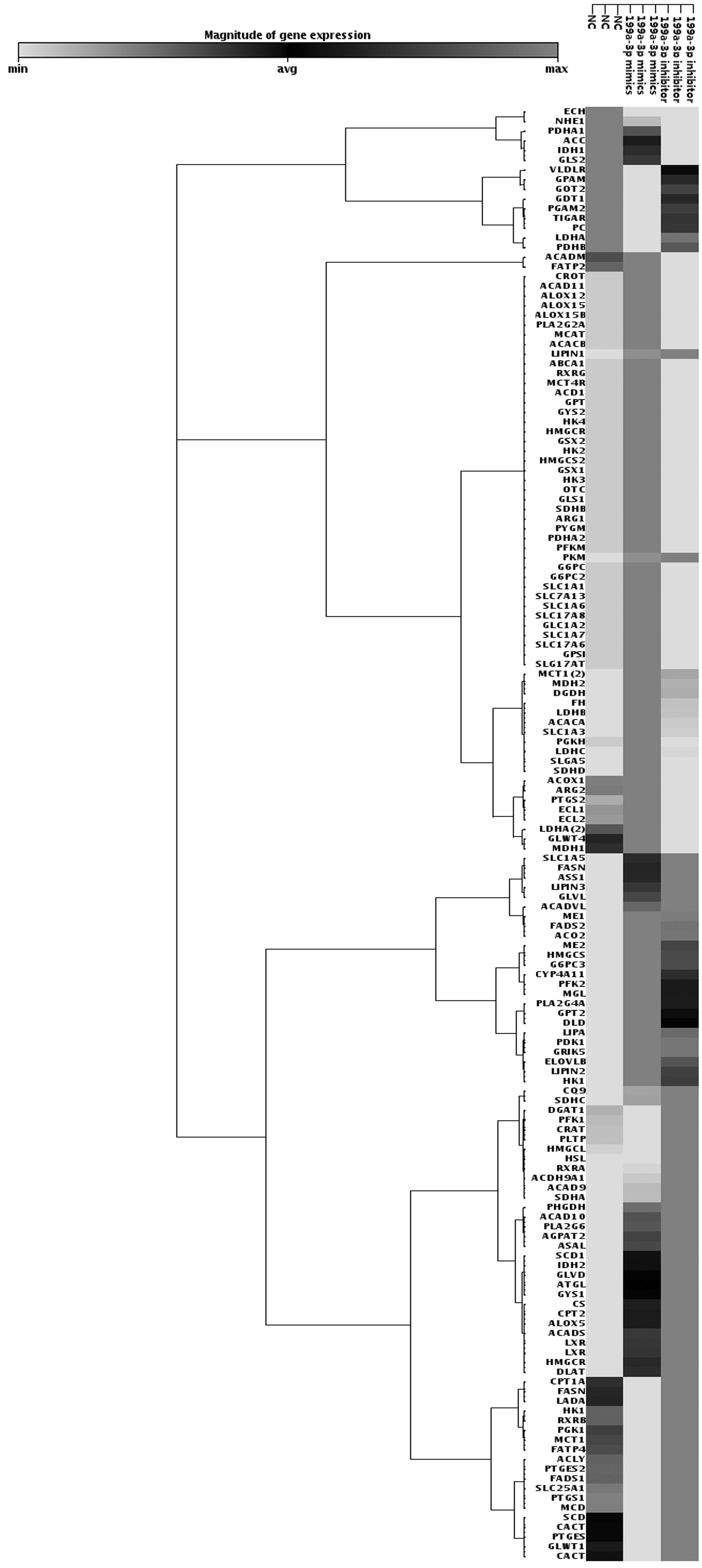
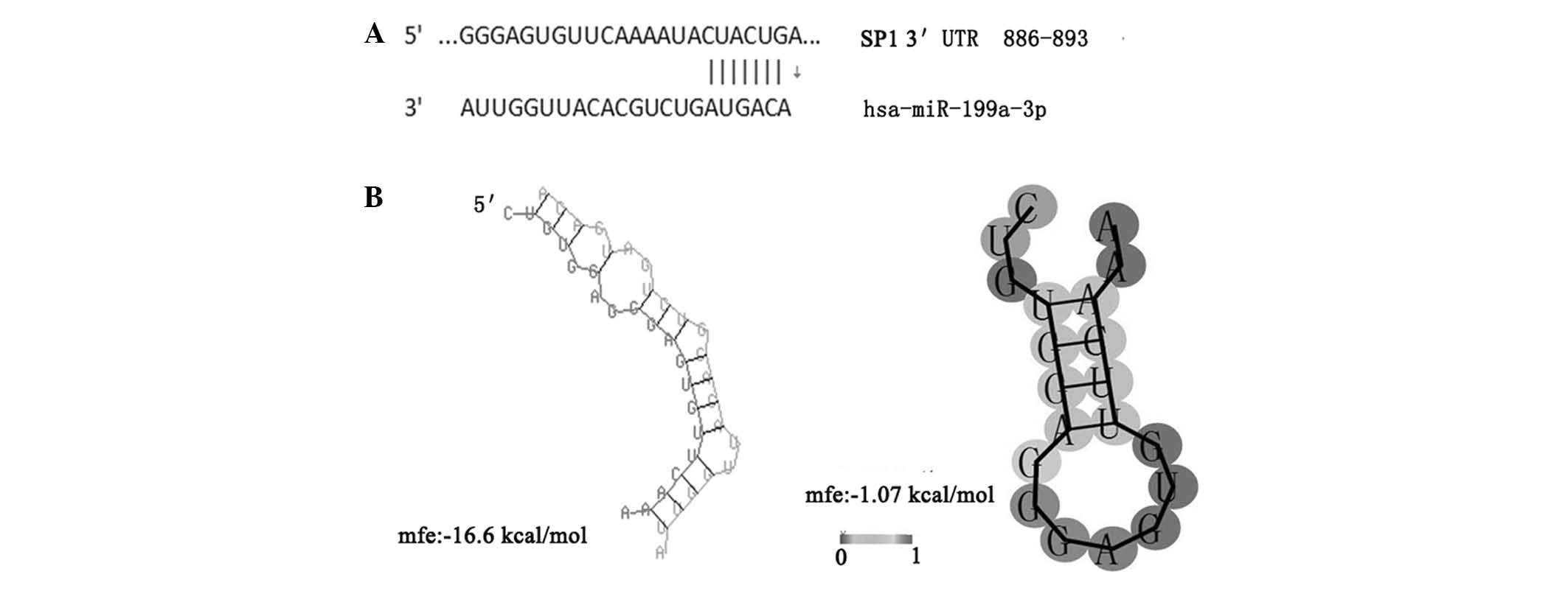
|
1
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lee RC, Feinbaum RL and Ambros V: The C.
elegans heterochronic gene lin-4 encoded small RNAs with antisense
complementarity to lin-14. Cell. 75:843–854. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Huang Y, Shen XJ, Zou Q, Wang SP, Tang SM
and Zhang GZ: Biological functions of microRNAs: A review. J
Physiol Biochem. 67:129–139. 2011. View Article : Google Scholar
|
|
5
|
Di Leva G and Croce CM: miRNA profiling of
cancer. Curr Opin Genet Dev. 23:3–111. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhang B, Pan X, Cobb GP and Anderson TA:
MicroRNAs as oncogenes and tumor suppressors. Dev Biol. 302:1–12.
2007. View Article : Google Scholar
|
|
7
|
Garzon R, Calin GA and Croce CM: MicroRNAs
in cancer. Annu Rev Med. 60:167–179. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Di Leva G and Croce CM: Roles of small
RNAs in tumor formation. Trends Mol Med. 16:257–267. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bezan A, Gerger A and Pichler M: MicroRNAs
in testicular cancer: Implications for pathogenesis, diagnosis,
prognosis and therapy. Anticancer Res. 34:2709–2713.
2014.PubMed/NCBI
|
|
10
|
Voorhoeve PM, le Sage C, Schrier M, Gillis
AJ, Stoop H, Nagel R, Liu YP, van Duijse J, Drost J, Griekspoor A,
et al: A genetic screen implicates miRNA-372 and miRNA-373 as
oncogenes in testicular germ cell tumors. Cell. 124:1169–1181.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chen BF, Gu S, Suen YK, Li L and Chan WY:
microRNA-199a-3p, DNMT3A, and aberrant DNA methylation in
testicular cancer. Epigenetics. 9:119–128. 2014. View Article : Google Scholar :
|
|
12
|
Rong Z, Li D and Liu X, Liu Z, Wu D and
Liu X: Screening for miRNAs and their potential targets in response
to TGF-β1 based on miRNA microarray and comparative proteomics
analyses in a mouse GC-1 spg germ cell line. Int J Mol Med.
35:821–828. 2015.PubMed/NCBI
|
|
13
|
Zhang Y, Fan KJ, Sun Q, Chen AZ, Shen WL,
Zhao ZH, Zheng XF and Yang X: Functional screening for miRNAs
targeting Smad4 identified miR-199a as a negative regulator of
TGF-β signalling pathway. Nucleic Acids Res. 40:9286–9297. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
15
|
Xiao L, Hu Z, Dong X, Tan Z, Li W, Tang M,
Chen L, Yang L, Tao Y, Jiang Y, et al: Targeting epstein-barr virus
oncoprotein LMP1-mediated glycolysis sensitizes nasopharyngeal
carcinoma to radiation therapy. Oncogene. 33:4568–4578. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Soga T: Cancer metabolism: Key players in
metabolic reprogramming. Cancer Sci. 104:275–281. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Han T, Kang D, Ji D, Wang X, Zhan W, Fu M,
Xin HB and Wang JB: How does cancer cell metabolism affect tumor
migration and invasion? Cell Adh Migr. 7:395–403. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chan B, Manley J, Lee J and Singh SR: The
emerging roles of microRNAs in cancer metabolism. Cancer Lett.
356:301–308. 2015. View Article : Google Scholar
|
|
19
|
Brenner B, Hoshen MB, Purim O, David MB,
Ashkenazi K, Marshak G, Kundel Y, Brenner R, Morgenstern S, Halpern
M, et al: MicroRNAs as a potential prognostic factor in gastric
cancer. World J Gastroenterol. 17:3976–3985. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wan D, He S, Xie B, Xu G, Gu W, Shen C, Hu
Y, Wang X, Zhi Q and Wang L: Aberrant expression of miR-199a-3p and
its clinical significance in colorectal cancers. Med Oncol.
30:3782013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Feber A, Xi L, Pennathur A, Gooding WE,
Bandla S, Wu M, Luketich JD, Godfrey TE and Litle VR: MicroRNA
prognostic signature for nodal metastases and survival in
esophageal adenocarcinoma. Ann Thorac Surg. 91:1523–1530. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Le Floch R, Chiche J, Marchiq I, Naiken T,
Ilc K, Murray CM, Critchlow SE, Roux D, Simon MP and Pouysségur J:
CD147 subunit of lactate/H+ symporters MCT1 and hypoxia-inducible
MCT4 is critical for energetics and growth of glycolytic tumors.
Proc Natl Acad Sci USA. 108:16663–16668. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Pinheiro C, Albergaria A, Paredes J, Sousa
B, Dufloth R, Vieira D, Schmitt F and Baltazar F: Monocarboxylate
transporter 1 is up-regulated in basal-like breast carcinoma.
Histopathology. 56:860–867. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Miranda-Gonçalves V, Honavar M, Pinheiro
C, Martinho O, Pires MM, Pinheiro C, Cordeiro M, Bebiano G, Costa
P, Palmeirim I, et al: Monocarboxylate transporters (MCTs) in
gliomas: Expression and exploitation as therapeutic targets. Neuro
Oncol. 15:172–188. 2013. View Article : Google Scholar :
|
|
25
|
Pinheiro C, Longatto-Filho A,
Azevedo-Silva J, Casal M, Schmitt FC and Baltazar F: Role of
monocarboxylate transporters in human cancers: State of the art. J
Bioenerg Biomembr. 44:127–139. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Walters DK, Arendt BK and Jelinek DF:
CD147 regulates the expression of MCT1 and lactate export in
multiple myeloma cells. Cell Cycle. 12:3175–3183. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Feng L, E LL, Soloveiv MM, Wang DS, Zhang
B, Dong YW and Liu HC: Synergistic cytotoxicity of cisplatin and
Taxol in overcoming Taxol resistance through the inhibition of LDHA
in oral squamous cell carcinoma. Oncol Lett. 9:1827–1832.
2015.PubMed/NCBI
|
|
28
|
Zhang D, Tai LK, Wong LL, Chiu LL, Sethi
SK and Koay ES: Proteomic study reveals that proteins involved in
metabolic and detoxification pathways are highly expressed in
her-2/neu-positive breast cancer. Mol Cell Proteomics. 4:1686–1696.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Chen G, Gharib TG, Wang H, Huang CC, Kuick
R, Thomas DG, Shedden KA, Misek DE, Taylor JM, Giordano TJ, et al:
Protein profiles associated with survival in lung adenocarcinoma.
Proc Natl Acad Sci USA. 100:13537–13542. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Duan Z, Lamendola D, Yusuf R, Penson R,
Preffer F and Seiden M: Overexpression of human phosphoglycerate
kinase 1 (PGK1) induces a multidrug resistance phenotype.
Anticancer Res. 22:1933–1941. 2002.PubMed/NCBI
|
|
31
|
Li L, He S, Sun JM and Davie JR: Gene
regulation by Sp1 and Sp3. Biochem Cell Biol. 82:460–471. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Solomon SS, Majumdar G, Martinez-Hernandez
A and Raghow R: A critical role of Sp1 transcription factor in
regulating gene expression in response to insulin and other
hormones. Life Sci. 83:305–312. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Safe S and Abdelrahim M: Sp transcription
factor family and its role in cancer. Eur J Cancer. 41:2438–2448.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Vizcaíno C, Mansilla S and Portugal J: Sp1
transcription factor: A long-standing target in cancer
chemotherapy. Pharmacol Ther. 152:111–124. 2015. View Article : Google Scholar : PubMed/NCBI
|