Introduction
The human interferon regulatory factor (IRF) family
comprises nine cellular IRFs, each of which has pleiotropic
biological functions (1). IRF5 is
important in the induction of type I interferons (IFNs),
proinflammatory cytokines and chemokines. Therefore, it is involved
in innate and adaptive immunity (2–5).
There is increasing evidence showing that IRF5 occupies a prominent
place among the genetic factors involved in the susceptibility to
human systemic lupus erythematosus (SLE), Sjögren's syndrome and
rheumatoid arthritis (6–8). Previous studies have demonstrated
that the expression, alternative splicing and protein levels of
IRF5 were significantly elevated in primary purified peripheral
blood mononuclear cells from patients with SLE (6,9).
As a significant factor mediating autoimmune
diseases, IRF5 exhibits certain tumour-suppressor properties, as it
can induce p21, B cell lymphoma 2-antagonist/killer 1, B cell
lymphoma 2-associated X protein and Caspase 8 (10–12).
The ectopic expression of IRF5 reduces the proliferation of chronic
myeloid leukaemia cells in vitro as a target of BCR-ABL
kinase (13). The expression of
IRF5 is reduced in gastric cancer, and is associated with
progression and metastasis of breast cancer (14,15).
By contrast, the tumour-promoting effect of IRF5 has also been
reported. IRF5 is expressed at high levels in primary and
immortalised thyroid carcinoma, but not in normal thyrocytes,
whereas ectopic expression of IRF5 increases the proliferation of
malignant thyroid cells (16). A
high expression level of IRF5 is specific to Hodgkin's lymphoma
cells and is crucial for their survival (17). Different functions of IRF5 have
been reported functions, as it exists as multiple alternatively
spliced variants, each with distinct cell type-specific expression
and function (18).
In addition, genetic polymorphisms in IRF5, which
lead to an increase in its expression, are associated with
inflammatory and autoimmune diseases. Rs2004640, the first
single-nucleotide polymorphism (SNP) to be identified, is
associated with the elevated expression of multiple isoforms of
IRF5, and is an important genetic risk factor for SLE (19). Another SNP (rs77571059, CGGGG
indel) is found 64 bp upstream of the transcription start site
(TSS) for exon 1A, which has four copies (4×) of the CGGGG repeat
sequence. The CGGGG 4× variant allows additional binding of the Sp1
transcription factor and is associated with the increased
expression of IRF5 (20). Sp1 was
the first among all the transcription factors to be identified and
cloned, and is responsible for the transcription of several
mammalian and viral genes with abundant GC boxes in their promoter
region through three C2H2-type zinc fingers in the C-terminal
domain (21). However, the
transcriptional regulation of IRF5, and whether Sp1 can regulate
the expression of IRF5 remains to be fully elucidated.
In the present study, it was determined that the
exogenous expression of Sp1 led to a significant increase in
promoter activity and the mRNA expression of IRF5. These findings
suggested that Sp1 positively regulated the transcription of IRF5
through binding to the minimal promoter region of IRF5 and
indicated a potential application of Sp1 as a target for treatment
of IRF5 related diseases.
Materials and methods
Cell culture and reagents
Human embryonic kidney (HEK) 293 and Hela cells were
obtained from American Type Culture Collection (Manassas, VA, USA).
The cells were maintained in Dulbecco's high glucose modified
Eagle's medium (DMEM) with 10% heat-inactivated fetal bovine serum
(FBS), supplemented with penicillin (100 U/ml) and streptomycin
(100 µg/ml). Mithramycin was purchased from Sangon Biotech
(Shanghai, China). Bovine serum albumin (BSA) was purchased from
Santa Cruz Biotechnology, Inc. (Dallas, TX, USA).
Plasmids and small interfering RNA
(siRNA)
The DNA sequence (−1,760 to +62) of the IRF5
promoter region was amplified by polymerase chain reaction (PCR)
and digested with KpnI and BglII (Thermo Fisher
Scientific, Inc., Waltham, MA, USA), followed by being subcloned
into the promoter-less luciferase expression plasmid, pGL3-Basic
(Promega Corp., Madison, WI, USA). The resulting plasmid was termed
pGL-1760/+62. Truncated plasmids of the IRF5 promoter were
constructed using pGL-1760/+62 as a template. The potential
transcription binding sites for Sp1 were identified using online
software TFSEARCH version 1.3 (www.cbrc.jp/research/db/TFSEARCH.html) and JASPAR
database version 5.0 (jaspar.genereg.net). A series of mutant plasmids of
pGL-179/+62 carrying a number of nucleotide substitutions
(Mut-Sp1-A, Mut-Sp1-B Mut-Sp1-C, Mut-Sp1-A+B, Mut-Sp1-A+C,
Mut-Sp1-B+C and Mut-Sp1-A+B+C) were constructed using a
site-directed mutagenesis kit (Takara Bio, Inc., Otsu, Japan). The
names of the plasmids and corresponding olignucleotides are shown
in Table I. The pN3-empty and
pN3-Sp1 expression plasmids were donated by Dr Guntram Suske
(University of Marburg, Marburg, Germany). The siRNAs were
synthesised and high performance purified (GenePharma, Shanghai,
China). The targeted sequence used to silence specific gene
transcription was sense 5′-AUCACUCCAUGGAUGAAAUGATT-3′ for Sp1. The
sequence of the control siRNA was 5′-UUCUCCGAACGUGUCACGUTT-3′.
 | Table IOligonucleotide sequences used for the
generation of reporter constructs. |
Table I
Oligonucleotide sequences used for the
generation of reporter constructs.
| Plasmid | Primer sequence
(5′-3′) |
|---|
| PGL-1760 | Sense:
5′-CGGGGTACCCTACCCATTCACATTTTCCCCATCC-3′ |
| Antisense:
5′-GGAAGATCTGGGACCAAGCTGAGCTCTGC-3′ |
| PGL-1080 | Sense:
5′-CGGGGTACCTACACCTGCTGCCTGTTGACCAAT-3′ |
| Antisense:
5′-GGAAGATCTGGGACCAAGCTGAGCTCTGC-3′ |
| PGL-503 |
Sense:5′-CGGGGTACCCAGGGTTTGAGGATGAGAAAGGCAC-3′ |
| Antisense:
5′-GGAAGATCTGGGACCAAGCTGAGCTCTGC-3′ |
| PGL-179 | Sense:
5′-CGGGGTACCAGGGCACCGCGCCGTCTGGCATCTC-3′ |
| Antisense:
5′-GGAAGATCTGGGACCAAGCTGAGCTCTGC-3′ |
| PGL-40 | Sense:
5′-CGGGGTACCAGCAGCAGCTGCCCAGGGGCGG-3′ |
| Antisense:
5′-GGAAGATCTGGGACCAAGCTGAGCTCTGC-3′ |
| PGL-9 | Sense:
5′-CGGGGTACCAGACGCGGAAGTGCCCGGCAG-3′ |
| Antisense:
5′-GGAAGATCTGGGACCAAGCTGAGCTCTGC-3′ |
| Mut-Sp1-A | Sense:
5′-ATTCGCGGTTTTGGGCGGGGCACTGCC-3′ |
| Antisense:
5′-CCACTCCGGGCCCCGCACTGACCTG-3′ |
| Mut-Sp1-B | Sense:
5′-GGGCGGGGTTTGGCACTGCCCGCGCCCGGAG-3′ |
| Antisense:
5′-CGCGAATCCACTCCGGGCCCCGCACTG-3′ |
| Mut-Sp1-C | Sense:
5′-TGCCCAGGTTTTGGGGCGGCAAGACGCGGAAG-3′ |
| Antisense:
5′-GCTGCTGCTGAGCTCCGGGCGCGGGCAG-3′ |
| Mut-Sp1-A+B | Sense: 5′
TTGGGTTTGGCACTGCCCGCGCCCGGAG-3′ |
| Antisense:
5′AACCGCGAATCCACTCCGGGCCCCGCAC-3′ |
| Mut-Sp1-A+C | Sense: 5′
ATTCGCGGTTTTGGGCGGGGCACTGCC-3′ |
| Antisense:
5′CCACTCCGGGCCCCGCACTGACCTG-3′ |
| Mut-Sp1-B+C | Sense:
5′GGTTTGGCACTGCCCGCGCCCGGAGCTCAG-3′ |
| Antisense:
5′CCGCCCCGCGAATCCACTCCGGGCCCCG-3′ |
| Mut-SP1-A+B+C | Sense: 5′
TTGGGTTTGGCACTGCCCGCGCCCGGAG-3′ |
| Antisense:
5′AACCGCGAATCCACTCCGGGCCCCGCAC-3′ |
Transient transfection and luciferase
assays
Transient transfection of the HEK 293 and Hela cells
was performed using Lipofectamine™ 3000 (Invitrogen; Thermo Fisher
Scientific, Inc.), according to the manufacturer's protocol. The
cells were seeded into 96-well plates (1.5×104/well) 24
h prior to transfection. For the Sp1 siRNA or Sp1 overexpression
experiments, reporter plasmids containing siRNA for Sp1 or the Sp1
expression plasmid were cotransfected into the cells and harvested
after 24 h. For the mithramycin experiment, 24 h following
transfection with the reporter plasmids, the cells were treated
with mithramycin (100 nM) or distilled water for 24 h to measure
luciferase activity. Luciferase activity was detected using the
Dual Reporter assay system (Promega Corp.). All experiments were
performed independently in triplicate.
Electrophoretic mobility shift analysis
(EMSA)
Nuclear extracts were obtained using nuclear and
cytoplasmic extraction reagents (Thermo Fisher Scientific, Inc.),
according to the manufacturer's protocol. As the Sp1-A and Sp1-B
binding sites overlapped, probe A comprised these two binding sites
(−85 to −48: 5′-AGTGGATTCGCGGGGCGGGGCGGGGCACTGCCCGCGC-3′) labeled
with biotin, and probe B contained the Sp1-C binding site (−36 to
−6: 5′-AGCAGCTGCCCAGGGGCGGGGGCGGCAAGA-3′). The nuclear protein
extract (10 µg) was incubated in binding buffer for 10 min
at room temperature, following which the biotinylated probes were
added, and the reaction mixture was incubated at room temperature
for another 20 min. For the competition experiments, unlabeled
probes were added to the reaction mixture at 100-fold excess molar
concentration. In the supershift assays, 1.5 µg of Sp1
antibody (cat. no. sc-59; 1:200; Santa Cruz Biotechnology, Inc.,
Santa Cruz, CA, USA) was preincubated for 20 min at room
temperature. The binding reactions were assessed by electrophoresis
through a 6% polyacrylamide gel, run for 1 h at 100 V, followed by
transfer onto a nylon membrane at 300 mA for 30 min, cross-linking
for 5 min under UV light, and detection with biotin-labeled DNA,
according to the protocol of the Light Shift Chemiluminescent EMSA
kit (Pierce Biotechnology, Inc., Rockford, IL, USA).
Chromatin immunoprecipitation (ChIP)
assay
A ChIP assay was performed using the EZ-Magna Chip™
A kit (cat. no. 17-408; EMD Millipore, Billerica, MA, USA)
according to the manufacturer's protocol. A total of
1×107 Hela cells were fixed in 1% formaldehyde at room
temperature for 10 min. The cell lysates were sonicated to generate
200–1,000 bp DNA fragments. The chromatin was immunoprecipitated
using anti-acetyl histone H3 antibody (from kit; 1:100; EMD
Millipore), anti-rabbit IgG antibody (from kit; 1:100; EMD
Millipore) and anti-Sp1 antibody (1:50; Santa Cruz Biotechnology,
Inc.). Following reverse cross-linking and DNA purification, DNA
from the input and immunoprecipitated samples was assayed using
semi-quantitative PCR with the following primers: Forward
5′-TGGCCCGAGGCTCAGCCCGGATCT-3′ and reverse
5′-TCCGCCAACCTGCCGGGCACTTCC-3′. The PCR products were analysed
using 2% agarose gel electrophoresis.
qPCR
Total RNA of the HEK 293 cells was extracted using
TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) and
then reverse transcribed use the PrimeScript RT Master Mix Perfect
Real Time kit (Takara Bio, Inc.). qPCR analysis was performed with
the Applied Biosystems Step One Plus Real-Time PCR system using
SYBR Premix Ex Taq (Takara Bio, Inc.), the following thermocycling
conditions were used: 95°C for 5 min; 40 cycles at 95°C for 15 sec,
60°C for 1 min. The specificity of amplification was assessed for
each sample by melting curve analysis. The expression of IRF5 was
normalised to GAPDH and the relative expression was calculated
using the comparative Cq method (22). The primers used were as follows:
IRF5, forward 5′-GGGCTTCAATGGGTCAACG-3′ and reverse
5′-GCCTTCGGTGTATTTCCCTG-3′, and GAPDH, forward
5′-TGGTATCGTGGAAGGACTCATGAC-3′ and reverse
5′-TGCCAGTGAGCTTCCCGTTCAGC-3′.
Statistical analysis
The results are expressed as the mean ± standard
error of the mean. Statistical analysis was performed using Student
t-test with SPSS software (version 20.0; IBM SPSS, Armonk,
NY, USA). P<0.05 was considered to indicate a statistically
significant difference.
Results
IRF5 minimal promoter is located in the
region between −179 and +62
To identify the IRF5 minimal promoter (Fig. 1A), a series of luciferase reporter
plasmids containing 5′ deletions of the putative (1,760/+62)
promoter region were cloned and transfected into Hela and HEK 293
cells (Fig. 1B and C). The
luciferase assays revealed a 20–22-fold increase in the promoter
activity of the pGL-1,760/+62, compared with the empty vector,
indicating a functional promoter in the −1,760/+62 region of IRF5.
Deletion of the sequence between −1,760 and −179 increased the
transcriptional activity by 1.6–2.0-fold, compared with
pGL-1,760/+62 (Fig. 1B),
suggesting negative regulatory elements were located in the
−1,760/−179 region. When the promoter was deleted to position −40,
transcriptional activity decreased significantly, by 69% in the
Hela cells and 73% in the HEK 293 cells, compared with pGL-179/+62,
which demonstrated that positive regulatory elements were located
in the −179/−40 region. Further 5′ deletion in the −9 to
+62-generated plasmid (pGL-9/+62) had a low level of promoter
activity, indicating that the minimal promoter of IRF5 was located
within the −179/+62 region relative to the TSS.
Sp1 is a transcriptional activator in the
minimal promoter region of IRF5
The sequence between −179 and −40 upstream of the
TSS had putative binding sites for Sp1, determined using online
software TFSEARCH version 1.3 and JASPAR database version 5.0
(Fig. 1A). To investigate the role
of these sites in the regulation of IRF5, a series of plasmids with
3–4 bp point mutations of Sp1 binding sites were constructed and
transiently transfected into Hela and HEK 293 cells. As shown in
Fig. 1C, mutations of Sp1-A, Sp1-B
and Sp1-C in the Hela cells reduced the promoter activity to 71, 68
and 37% of the pGL3-179/+62 promoter activity, respectively.
Similarly, mutation of Sp1-A, Sp1-B and Sp1-C in in HEK 293 cells
decreased the promoter activity to 62, 60 and 33% of the
pGL3-179/+62 promoter, respectively. However, combinatorial
mutations (double mutations), had minimal additional effect on
basal promoter activity. The triple mutation of Sp1-A+B+C reduced
the promoter activity more markedly. To determine whether Sp1
directly regulated the transcription of IRF5, an Sp1-overexpression
plasmid was introduced into Hela and HEK 293 cells. As shown in
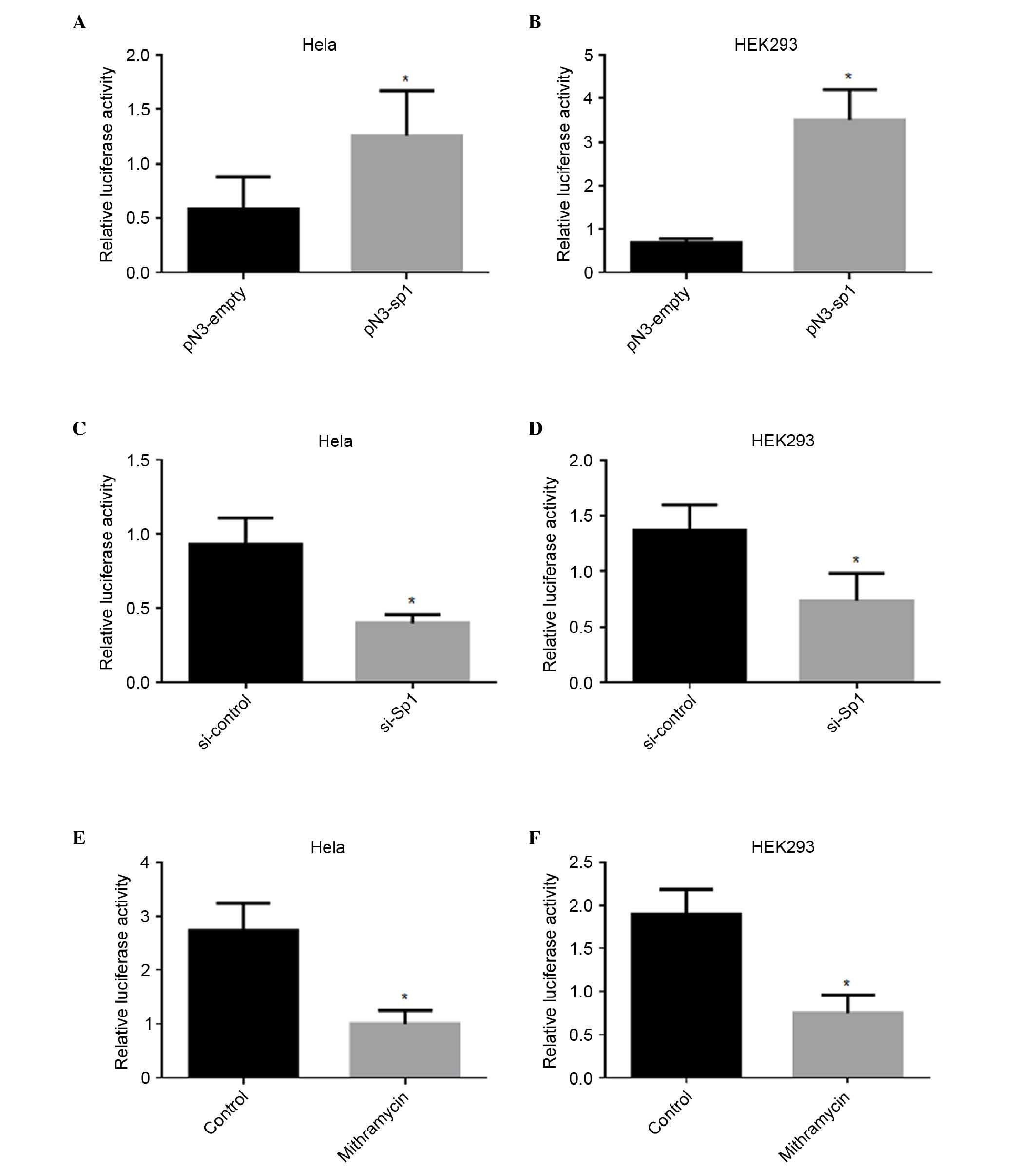
Fig. 2A and B, the overexpression
of Sp1 enhanced the luciferase activity of pGL3-179/+62 by 1-fold
in the Hela cells and 4-fold in the HEK293 cells, respectively.
Silencing of Sp1 reduces IRF5 promoter
activity
To further confirm the roles of Sp1 in the
regulation of IRF5 promoter activity, the pGL-179/+62 plasmid was
cotransfected together with Sp1 siRNA or si-control. As shown in
Fig. 2C and D, repression of the
endogenous expression of Sp1 induced a marked decrease in promoter
activity, by 57% in the Hela cells and 46% in the HEK 293 cells,
respectively. Mithramycin is known to bind to the GC box and
inhibit Sp1 binding (23).
Treatment with mithramycin also led to a significant decrease in
promoter activity, by 63% in the Hela cells and 61% in the HEK 293
cells, respectively (Fig. 2E and
F).
Sp1 binds to the minimal promoter of IRF5
in vitro and in vivo
To validate whether Sp1 bound to the IRF5 promoter
in vitro, an EMSA was performed. As shown in Fig. 3A and B, the nuclear extracts from
the Hela cells bound to the labeled wild-type probe A and probe B,
and formed shifted bands (lane 2). No shifted bands were observed
when bovine serum albumin was substituted for the nuclear extracts
(lane 1). The shifted bands were eliminated when incubated with
100-fold excess unlabeled probe (lane 3). As shown in the lane 4,
super-shifted bands were produced when incubated with Sp1 antibody,
and the density of the corresponding shifted bands were reduced,
indicating that Sp1 bound to multiple motifs in the IRF5 promoter
in vitro. In addition, a ChIP assay was performed to examine
whether Sp1 interacted with the IRF5 promoter in vivo. The
cross-linked chromatin prepared from the Hela cells was
immunoprecipitated with anti-acetyl histone H3, anti-IgG and
anti-Sp1 antibodies. The DNA precipitated in the complexes was then
subjected to PCR with primers covering the regions of the IRF5
promoter sequence. As shown in Fig.
3C, the anti-acetyl histone H3 and anti-Sp1 antibodies
immunoprecipitated the sequence of the IRF5 promoter, whereas the
non-specific IgG antibody failed to immunoprecipitate this
sequence, suggesting that Sp1 was able to bind to the minimal
promoter of IRF5 in vivo.
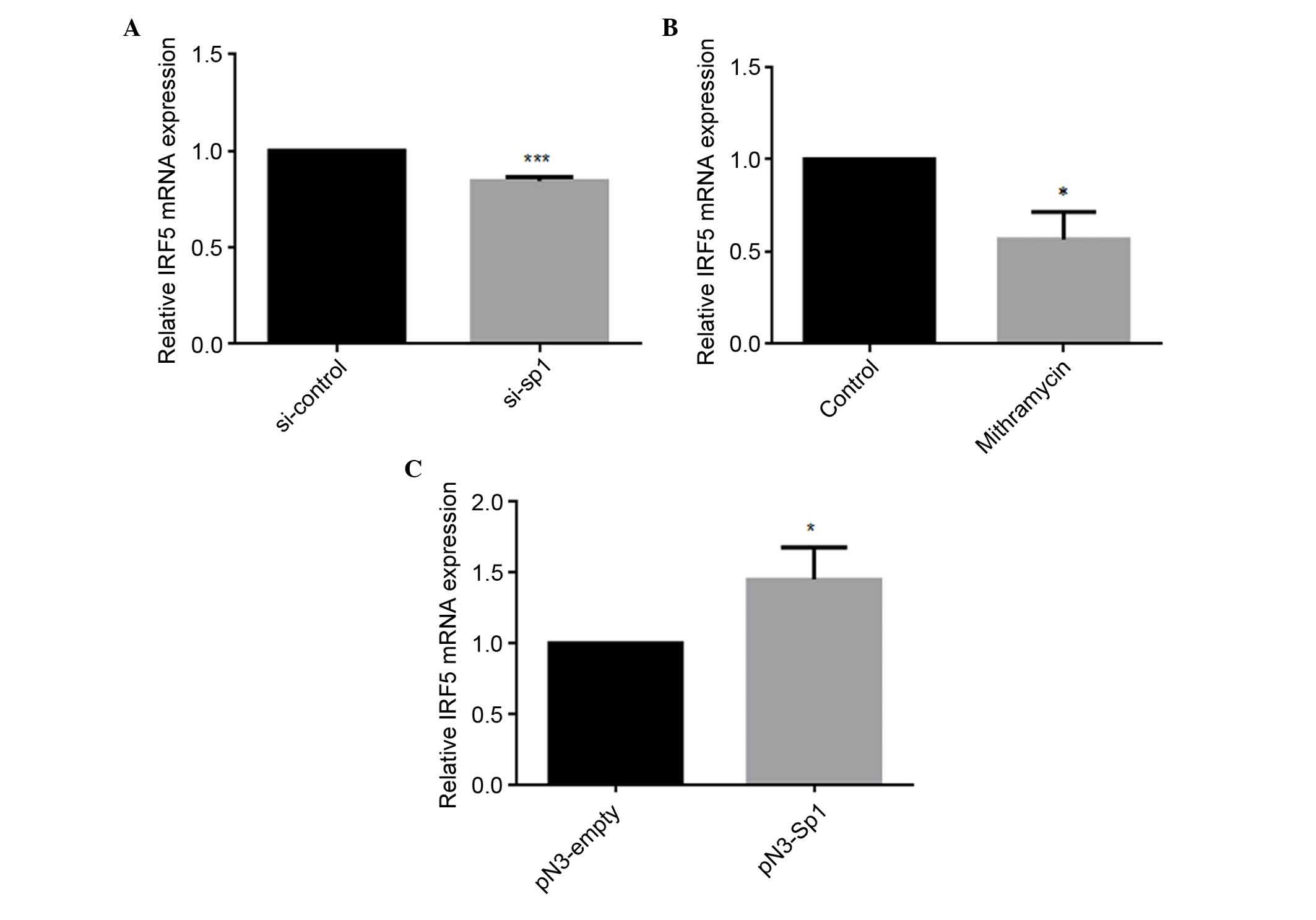
Sp1 regulates the mRNA expression level
of IRF5
To further examine the role of Sp1 in regulating the
expression of IRF5, siRNA, mithramycin or the Sp1-overexpression
vector were transfected into HEK293 cells. The mRNA levels of IRF5
were detected using qPCR analysis. As shown in Fig. 4A and B, siRNA and mithramycin
reduced the mRNA levels of IRF5 by 16 and 44%, respectively. By
contrast, the overexpression of Sp1 resulted in a 1.45-fold
increase in the mRNA expression of IRF5, compared with the
pN3-empty negative control (Fig.
4C), indicating that Sp1 upregulated the mRNA expression level
of IRF5.
Discussion
In our previous studies, cloning of IRF3 and its
spliced variant promoters was performed, and it was identified that
Sp1/Sp3 positively regulates the transcription of IRF3 through
binding to the Sp1/NRF-1 binding site in the core promoter region
of IRF3 (24,25). The nine distinct alternatively
spliced human IRF5 mRNAs (V1-V9) were previously identified by
Mancl et al (18), who
showed that V1 and V3 had distinct TSSs, and these two discrete
promoters were cloned. V5 and V6 were expressed in peripheral blood
mononuclear cells, whereas V7, V8 and V9 were expressed only in
human cancer cells (18). In
addition, Clark et al (26)
revealed that IRF5 has four promoters for its four first exons: 1A,
1B, 1C and 1D. These promoters were all found to contain Sp1
putative binding sites. When treated with imiquimod, a Toll-like
receptor 7 ligand, the mRNA levels of IRF5 were doubled, and the 1A
and 1D promoter activities were increased.
In the present study, the 5′-flanking region of the
wild-type, IRF5, which had the most integrated reference sequence
(V5), was cloned, which was located between the V1 and V3 promoter.
The pGL-1760/+62 construct contained the 1A and 1D promoter without
the 1B or 1C promoter, however, the pGL-179/+62 construct contained
only the 1A promoter, which possibly included the 4× variant of the
CGGGG indel, but no Rs2004640. When subcloned into a luciferase
reporter vector and transfected into Hela and HEK 293 cells, the
5′-flanking region showed promoter activity, and the minimal
promoter was demonstrated to be located within the −179 to +62
region. Bioinformatics analyses suggested that the minimal promoter
contained several canonical Sp1 binding sites. Accumulating
evidence shows that Sp1 is critical for the transcriptional
initiation of TATA-less promoters (27,28).
Currently, the Sp family consists of nine distinct proteins (Sp1-9)
(29), among which Sp1, Sp2, Sp3
and Sp4 comprise two major glutamine-rich transactivation domains,
which are essential for transcriptional activation (28). They bind to GC/GT boxes via three
C2H2-type zinc fingers to regulate the expression of housekeeping,
tissue-specific and viral genes and are involved in several
important aspects of cellular function (30). In the present study, three Sp1
binding sites were identified within the minimal promoter of IRF5.
Of note, all the putative Sp1 sites were structurally identical to
the consensus binding site (G/T)GGGCGG(G/A)(G/A) (C/T) (31), although Sp-C was located in the
reverse strand.
Mutational analyses of the putative Sp1 sites
revealed that individual mutations of the Sp1-A, Sp1-B or Sp1-C
sites reduced the activity of the promoter, and the most marked
reduction was found in combinatorial mutation of the Sp1-A, Sp1-B
and Sp1-C sites. The EMSA and ChIP experiments demonstrated that
Sp1 was able to bind to the promoter of IRF5 in vitro and
vivo, respectively. In addition, the overexpression of Sp1
increased promoter activity and the endogenous mRNA level of IRF5.
The present study also identified that knockdown of Sp1 using siRNA
technology resulted in a significant decrease in the
transcriptional activity of the IRF5 promoter and the mRNA level of
IRF5. The present study also showed that treatment of the Hela and
HEK 293 cells with mithramycin led to a reduction in the promoter
activity and mRNA expression of IRF5. Taken together, these results
indicated that Sp1 was essential in the basal transcriptional
regulation of IRF5.
Sp1 and Sp3 have the same binding sites, however,
whether Sp3 regulates the expression of IRF5 remains to be
elucidated. Post-translational modifications of Sp1 and
interactions with other transcription factors can also affect
transcriptional activity, therefore, the further investigation of
the underlying regulatory mechanism is required.
In conclusion, the present study found that the
primary activating regulatory region of human IRF5 was located in
its minimal promoter region between nucleotides −179 and +62. In
addition, it was shown that Sp1 was able to bind to the multiple
sites in IRF5 promoter region, and was involved in the
transcriptional regulation of IRF5 at the basal level. These
results may provide novel insight into the molecular mechanisms
underlying the regulation of IRF5.
Abbreviations:
|
BSA
|
bovine serum albumin
|
|
ChIP
|
chromatin immunoprecipitation
|
|
DMEM
|
Dulbecco's modified Eagle's medium
|
|
EMSA
|
electrophoretic mobility shift
assay
|
|
FBS
|
fetal bovine serum
|
|
IRF5
|
interferon regulatory factor 5
|
|
siRNA
|
small interfering RNA
|
|
SLE
|
systemic lupus erythematosus
|
|
SNP
|
single-nucleotide polymorphism
|
|
Sp1
|
specificity protein 1
|
|
TSS
|
transcription start site
|
Acknowledgments
The authors would like to thank Dr G. Suske for the
Sp1 expression plasmids. This study was supported by the National
Natural Science Foundation of China (grant nos. 81170661 to G.P.Z.,
81300023 to R.J. and 81302531 to Hua-Guo Xu), the Natural Science
Foundation of Jiangsu Province of China (grant nos. BK20131018 to
H.G.X. and BK20131020 to R.J.), the Specialized Research Fund for
the Doctoral Program of Higher Education (grant no. 20113234110010
to G.P.Z.), and the Project Funded by the Priority Academic Program
Development of Jiangsu Higher Education Institutions.
References
|
1
|
Savitsky D, Tamura T, Yanai H and
Taniguchi T: Regulation of immunity and oncogenesis by the IRF
transcription factor family. Cancer Immunol Immunother. 59:489–510.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Takaoka A, Yanai H, Kondo S, Duncan G,
Negishi H, Mizutani T, Kano S, Honda K, Ohba Y, Mak TW and
Taniguchi T: Integral role of IRF-5 in the gene induction programme
activated by Toll-like receptors. Nature. 434:243–249. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yanai H, Chen HM, Inuzuka T, Kondo S, Mak
TW, Takaoka A, Honda K and Taniguchi T: Role of IFN regulatory
factor 5 transcription factor in antiviral immunity and tumor
suppression. Proc Natl Acad Sci USA. 104:3402–3407. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Krausgruber T, Saliba D, Ryzhakov G,
Lanfrancotti A, Blazek K and Udalova IA: IRF5 is required for
late-phase TNF secretion by human dendritic cells. Blood.
115:4421–4430. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Barnes BJ, Kellum MJ, Field AE and Pitha
PM: Multiple regulatory domains of IRF-5 control activation,
cellular localization, and induction of chemokines that mediate
recruitment of T lymphocytes. Mol Cell Biol. 22:5721–5740. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Feng D, Stone RC, Eloranta ML,
Sangster-Guity N, Nordmark G, Sigurdsson S, Wang C, Alm G, Syvänen
AC, Rönnblom L and Barnes BJ: Genetic variants and
disease-associated factors contribute to enhanced interferon
regulatory factor 5 expression in blood cells of patients with
systemic lupus erythematosus. Arthritis Rheum. 62:562–573.
2010.PubMed/NCBI
|
|
7
|
Kim K, Cho SK, Han TU, Kim JH, Kang SJ,
Kang C and Bae SC: A redundant epistatic interaction between IRF5
and STAT4 of the type I interferon pathway in susceptibility to
lupus and rheumatoid arthritis. Lupus. 22:1336–1340. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lessard CJ, Li H, Adrianto I, Ice JA,
Rasmussen A, Grundahl KM, Kelly JA, Dozmorov MG, Miceli-Richard C,
Bowman S, et al: Variants at multiple loci implicated in both
innate and adaptive immune responses are associated with Sjögren's
syndrome. Nat Genet. 45:1284–1292. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Stone RC, Feng D, Deng J, Singh S, Yang L,
Fitzgerald-Bocarsly P, Eloranta ML, Rönnblom L and Barnes BJ:
Interferon regulatory factor 5 activation in monocytes of systemic
lupus erythematosus patients is triggered by circulating
autoantigens independent of type I interferons. Arthritis Rheum.
64:788–798. 2012. View Article : Google Scholar :
|
|
10
|
Barnes BJ, Kellum MJ, Pinder KE, Frisancho
JA and Pitha PM: Interferon regulatory factor 5, a novel mediator
of cell cycle arrest and cell death. Cancer Res. 63:6424–6431.
2003.PubMed/NCBI
|
|
11
|
Bi X, Feng D, Korczeniewska J, Alper N, Hu
G and Barnes BJ: Deletion of Irf5 protects hematopoietic stem cells
from DNA damage-induced apoptosis and suppresses
γ-irradiation-induced thymic lymphomagenesis. Oncogene.
33:3288–3297. 2014. View Article : Google Scholar
|
|
12
|
Hu G and Barnes BJ: IRF-5 is a mediator of
the death receptor-induced apoptotic signaling pathway. J Biol
Chem. 284:2767–2777. 2009. View Article : Google Scholar
|
|
13
|
Massimino M, Consoli ML, Mesuraca M,
Stagno F, Tirrò E, Stella S, Pennisi MS, Romano C, Buffa P, Bond
HM, et al: IRF5 is a target of BCR-ABL kinase activity and reduces
CML cell proliferation. Carcinogenesis. 35:1132–1143. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yamashita M, Toyota M, Suzuki H, Nojima M,
Yamamoto E, Kamimae S, Watanabe Y, Kai M, Akashi H, Maruyama R, et
al: DNA methylation of interferon regulatory factors in gastric
cancer and noncancerous gastric mucosae. Cancer Sci. 101:1708–1716.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bi X, Hameed M, Mirani N, Pimenta EM,
Anari J and Barnes BJ: Loss of interferon regulatory factor 5
(IRF5) expression in human ductal carcinoma correlates with disease
stage and contributes to metastasis. Breast Cancer Res.
13:R1112011. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Massimino M, Vigneri P, Fallica M, Fidilio
A, Aloisi A, Frasca F and Manzella L: IRF5 promotes the
proliferation of human thyroid cancer cells. Mol Cancer. 11:212012.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kreher S, Bouhlel MA, Cauchy P, Lamprecht
B, Li S, Grau M, Hummel F, Köchert K, Anagnostopoulos I, Jöhrens K,
et al: Mapping of transcription factor motifs in active chromatin
identifies IRF5 as key regulator in classical Hodgkin lymphoma.
Proc Natl Acad Sci USA. 111:E4513–E4522. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Mancl ME, Hu G, Sangster-Guity N,
Olshalsky SL, Hoops K, Fitzgerald-Bocarsly P, Pitha PM, Pinder K
and Barnes BJ: Two discrete promoters regulate the alternatively
spliced human interferon regulatory factor-5 isoforms. Multiple
isoforms with distinct cell type-specific expression, localization,
regulation, and function. J Biol Chem. 280:21078–21090. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Graham RR, Kozyrev SV, Baechler EC, Reddy
MV, Plenge RM, Bauer JW, Ortmann WA, Koeuth T, González Escribano
MF; Argentine and Spanish Collaborative Groups; et al: A common
haplotype of interferon regulatory factor 5 (IRF5) regulates
splicing and expression and is associated with increased risk of
systemic lupus erythematosus. Nat Genet. 38:550–555. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Sigurdsson S, Göring HH, Kristjansdottir
G, Milani L, Nordmark G, Sandling JK, Eloranta ML, Feng D,
Sangster-Guity N, Gunnarsson I, et al: Comprehensive evaluation of
the genetic variants of interferon regulatory factor 5 (IRF5)
reveals a novel 5 bp length polymorphism as strong risk factor for
systemic lupus erythematosus. Hum Mol Genet. 17:872–881. 2008.
View Article : Google Scholar
|
|
21
|
Briggs MR, Kadonaga JT, Bell SP and Tjian
R: Purification and biochemical characterization of the
promoter-specific transcription factor, Sp1. Science. 234:47–52.
1986. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
23
|
Seznec J, Silkenstedt B and Naumann U:
Therapeutic effects of the Sp1 inhibitor mithramycin A in
glioblastoma. J Neurooncol. 101:365–377. 2011. View Article : Google Scholar
|
|
24
|
Xu HG, Jin R, Ren W, Zou L, Wang Y and
Zhou GP: Transcription factors Sp1 and Sp3 regulate basal
transcription of the human IRF-3 gene. Biochimie. 94:1390–1397.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ren W, Zhu LH, Xu HG, Jin R and Zhou GP:
Characterization of a spliced variant of human IRF-3 promoter and
its regulation by the transcription factor Sp1. Mol Biol Rep.
39:6987–6993. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Clark DN, Read RD, Mayhew V, Petersen SC,
Argueta LB, Stutz LA, Till RE, Bergsten SM, Robinson BS, Baumann
DG, et al: Four promoters of IRF5 respond distinctly to stimuli and
are affected by autoimmune-risk polymorphisms. Front Immunol.
4:3602013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kadonaga JT, Carner KR, Masiarz FR and
Tjian R: Isolation of cDNA encoding transcription factor Sp1 and
functional analysis of the DNA binding domain. Cell. 51:1079–1090.
1987. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Suske G: The Sp-family of transcription
factors. Gene. 238:291–300. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Suske G, Bruford E and Philipsen S:
Mammalian SP/KLF transcription factors: Bring in the family.
Genomics. 85:551–556. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Philipsen S and Suske G: A tale of three
fingers: The family of mammalian Sp/XKLF transcription factors.
Nucleic Acids Res. 27:2991–3000. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Tan NY and Khachigian LM: Sp1
phosphorylation and its regulation of gene transcription. Mol Cell
Biol. 29:2483–2488. 2009. View Article : Google Scholar : PubMed/NCBI
|