Introduction
Breast cancer (BC) is a leading cause of mortality
among women with, on average, 400,000 mortalities per year
(1). The incidence of BC has been
increased in Asian countries (2),
including Pakistan [the estimated rate is 38.4%, and every ninth
woman is at risk of the disease at certain stages in her life
(3)]. BC is caused by complex
inherited and environmental factors, and is therefore called a
multifactorial disease (4).
However, genetics of an individual play a vital role in the
development of BC. Tumor suppressor genes, including breast cancer
1 (BRCA1), BRCA2, phosphatase and tensin homolog (ʻPTENʼ) and tumor
protein 53, have been recognized in inherited BC syndromes.
Furthermore, DNA-repair genes, such as checkpoint kinase 2, ataxia
telangiectasia mutated (ʻATMʼ), BRCA1-interacting protein and
partner and localizer of BRCA2, have been moderately associated
with the risk of BC (5). However,
all these known common genes account for only 25% of the familial
BC cases (5), suggesting that an
additive effect of multiple susceptibility alleles with low
penetrance may be, in part, responsible for the risk of BC
(6). This hypothesis leads to a
polygenic model of susceptibility of BC to genetic factors, in
which a large number of low-risk variants having high frequencies
in populations may determine the overall risk of disease due to
their multiplicative effect (7).
Single nucleotide polymorphisms (SNPs) are the genetic factors that
have been considered as the key variations leading to BC
susceptibility among individuals (8). To date, >40 common low-risk
variants have been reported by genome-wide association studies
(GWASs) that are associated with BC risk (9). Certain studies have shown that the BC
risk is associated with different SNPs in three novel genes: Trinuc
leotide-repeat-containing 9 (TNRC9), fibroblast growth factor
receptor 2 (FGFR2) and mitogen-activated protein kinase kinase
kinase 1 (MAP3K1). It has been previously shown in numerous studies
that SNPs in the genes FGFR2, TNRC9 and MAP3K1 [or MEKK1 (MEK
kinase 1)] are associated with a high risk of BC in the general
population, as well as in BRCA2 mutation carriers (10–12).
FGFR2 is a tumor suppressor gene (located on
chromosome 10q26 with 22 exons), which is responsible for 10–15% of
breast tumors due to its overexpression (13,14).
Common SNPs and numerous mutations within, or neighboring, the
FGFR2 gene have been associated with the susceptibility of BC. For
example, two intronic polymorphic variants of the FGFR2 gene
(rs1219648 and rs2981582, which are located in intron 2) have been
closely associated with BC (10,15).
In another study, the SNP, rs1219648, was closely linked with the
early onset of sporadic BC in African, American and Chinese women,
specifically in young women (16),
the Azeri population of Iran (17)
and the North Indian population (18). GWASs also identified TNRC9 (located
on chromosome 16q12.1) as a BC susceptibility locus, and placed it
in one of the low-risk BC variants (15). Overexpression of the TNRC9 gene is
associated with a poor diagnosis, as it increases BC cell
propagation, migration and survival (19). The SNPs rs3803662 and rs8051542,
belonging to the TNRC9 gene, have been shown to be clearly
associated with BC in women of different ethnicities (10,20–22).
However, repeated studies among European, African-American and East
Asian individuals reported contradictory results (23–25).
MAP3K1 belongs to the MAPK family, and it exerts a pivotal role in
multiple normal and tumor cell types through being involved in
functions such as apoptosis, cell survival and cell
motility/migration (26).
Differential expression of MAP3KI has been reported in all BC
subtypes (7,10). The variant of rs889312 (MAP3K1) has
been demonstrated to be a powerful risk factor for the development
of BC in European and Asian ancestry populations (27,28).
However, similar SNPs do not show any association with the risk of
BC in women of African ancestry (29–31).
Taken together, the novel genetic variants of the genes FGFR2,
MAP3K1 and TNRC9 have shown a marked association with BC in
populations of diverse ethnicity (32), and the variants identified as BC
risk factors have revealed a variable impact on the risk of BC
associated with different populations. Hence, the replications of
previously BC associated loci in multiple populations are required
to explore the genetic heterogeneity of BC (18).
Therefore, the genetic association of five genetic
variants of the three genes, including FGFR2 (SNPs: rs1219648,
rs2981582), TNRC9 [new name: TOX high-mobility group box family
member 3 (TOX3), Ser51 variant; SNPs: rs8051542, rs380662), and
MAP3K1 (SNP: rs889312) was performed in female patients with BC, as
well as age-matched healthy subjects of Pakistan. Furthermore, the
association of variants with a BC risk for stratified groups of
patients, based on their clinicopathological characteristics, was
also determined.
Materials and methods
Sample collection
Blood samples of 100 patients with BC were collected
in k3 EDTA vials obtained from the Institute of Nuclear and
Medicine Oncology (INMOL), Lahore, and ITTEFAQ Hospital, Lahore.
Clinical data of the patients, including their estrogen receptor
(ER) and progester one receptor (PR) and human epidermal growth
factor receptor 2 (HER2) status, as well as patients with a family
history of BC, were collected from the files of patients. Informed
consent according to the declaration of Helsinki was obtained from
patients or from their relatives, as well as healthy women who were
involved in the study, and the COMSATS Ethics Committee approved
the study.
Extraction of DNA
DNA was extracted from the fresh blood samples of
patients with BC and healthy controls using either a DNA extraction
kit (K0512-Thermo Scientific Genomic DNA Purification kit; Thermo
Fisher Scientific, Waltham, MA, USA) or an organic method of DNA
extraction, as previously described (33). The DNA quality and quantity was
determined using agarose gel electrophoresis, as described
below.
Design of primers
The allele-specific amplification primers for
allelic variants of FGFR2, TNRC9 and MAP3K1 were designed using
Primer 3 (v. 0.4.0) software from the website, http://frodo.wi.mit.edu. Genomic DNA flanking the SNP
was amplified with allele-specific primers. Two different pairs of
primers were used for SNP amplification: One wild-type
allele-specific primer, and the other mutant allele-specific
primer. The universal primer was non-allele-specific, and identical
in wild and mutant genotypes of each marker. The protocol of
Hirotsu et al (34) was
followed for the design of the allele-specific primers. The primer
sequences, along with the optimized annealing temperatures for each
variant, are shown in Table I.
 | Table IList of primers for the
allele-specific amplification of the SNP regions of the genes,
FGFR2, MAP3K1 and TNRC9. |
Table I
List of primers for the
allele-specific amplification of the SNP regions of the genes,
FGFR2, MAP3K1 and TNRC9.
| Allele name | Name of primer | Primer sequence
(5′-3′) | Annealing
temperature |
|---|
| MAP3K1
rs889312 | Universal
forward |
5′-GACACAGGCATCAATTATTTCT-3′ | 57 |
| C reverse |
5′-GTAGTCTCTTAATTTGCACATG-3′ | 57 |
| A reverse |
5′-GTAGTCTCTTAATTTGCACATT-3′ | 57 |
| FGFR2
rs1219648 | Universal
forward |
5′-CATGATGTGGCCAAAGTCCA-3′ | 58 |
| A reverse |
5′-CATGGCCATCCTTGAAGAGT-3′ | 58 |
| G reverse |
5′-CATGGCCATCCTTGAAGAGC-3′ | 58 |
| FGFR2
rs2981582 | T forward |
5′-GCCACTTAATGAACCTGTTTGT-3′ | 56 |
| C forward |
5′-GCCACTTAATGAACCTGTTTGC-3′ | 56 |
| Universal
reverse |
5′-ACGCAACCCTCCTTCCTAAAC-3′ | 56 |
| TNRC9
rs8051542 | Universal
forward |
5′-GCCAGAAGTTTTCCATCTCT-3′ | 54 |
| T reverse |
5′-CTCCAATCATAGTGCTGCA-3′ | 54 |
| C reverse |
5′-CTCCAATCATAGTGCTGCG-3′ | 54 |
| TNRC9
rs3803662 | T forward |
5′-TTAATGCCTCTATAGCTGTCT-3′ | 53 |
| C forward |
5′-TTAATGCCTCTATAGCTGTCC-3′ | 53 |
| Universal
reverse |
5′-AGGAGACAAAGGTAGTAATGG-3′ | 53 |
Allele-specific polymerase chain reaction
(PCR) amplification
Allele-specific PCR was performed in a 20 µl
reaction volume containing 10 ng genomic DNA, 0.4 pM each
oligonucleotide primer, 1X PCR buffer, 200 µM
deoxynucleoside triphosphates, 2 mM MgCl2 and 2 U
Taq polymerase (all obtained from Thermo Fisher Scientific,
Inc.). The reactions were performed using the following PCR cycling
conditions: 3 min at 95°C for one cycle, 35 cycles at 95°C for 30
sec, with the different annealing temperatures as shown in Table I for 30 sec and 72°C for 30 sec,
followed by one cycle at 72°C for 7 min.
Agarose gel electrophoresis
Amplified products of SNPs were electrophoresed (80
V) on a 2.5% agarose gel stained with ethidium bromide and
visualized on UV trans-illuminator. The allelic variants of FGFR2,
TNRC9 and MAP3K1 were genotyped using a gel-based method, as
detailed below.
Sanger sequencing
In order to validate the gel-based method of SNP
genotyping, Sanger sequencing of purified PCR products of selected
samples was performed to confirm the different allelic variants of
FGFR2, TNRC9 and MAP3K1. Sequencing of the purified products using
universal primer (either reverse or forward, as shown in Table I) was performed with a Big Dye
Sequencing kit, according to the manufacturer's protocol (Applied
Biosystems Life Technologies, Foster City, CA, USA). The sequencing
chromatograms were analyzed using Genious software (version R9.1;
www.genious.com).
Statistical analysis
For estimating the association of genetic variants
with BC, the Chi-squared test was used, and the odds ratio (OR) and
95% confidence intervals (CIs) were also calculated. Fisher's exact
test was performed for determining the association of haplotypes
with the risk of BC. Statistical analysis was performed using
SHEsis online software (35), with
the exception of the Chi-squared test to compare the distribution
of SNP genotypes between the cancer group and healthy controls,
where SPSS version 17 (SPSS Inc., Chicago, IL, USA) was used.
P<0.05 was considered to indicate a statistically significant
value.
Results
The allele-specific amplification of five selected
low-risk variants of three genes, including FGFR2 (SNPs: rs2981582,
rs1219648), TNRC9 (SNPs: rs8051542, rs3803662), and MAP3K1 (SNP:
rs889312), revealed the presence of DNA fragments of 223, 177, 242,
210 and 219 bp in length, respectively, on 2% agarose gel
electrophoresis (Fig. 1). The
sequencing of selected PCR products for each genetic marker
revealed 100% concordance with the gel electrophoretic method. The
sequencing chromatograms for the two allelic variants of each
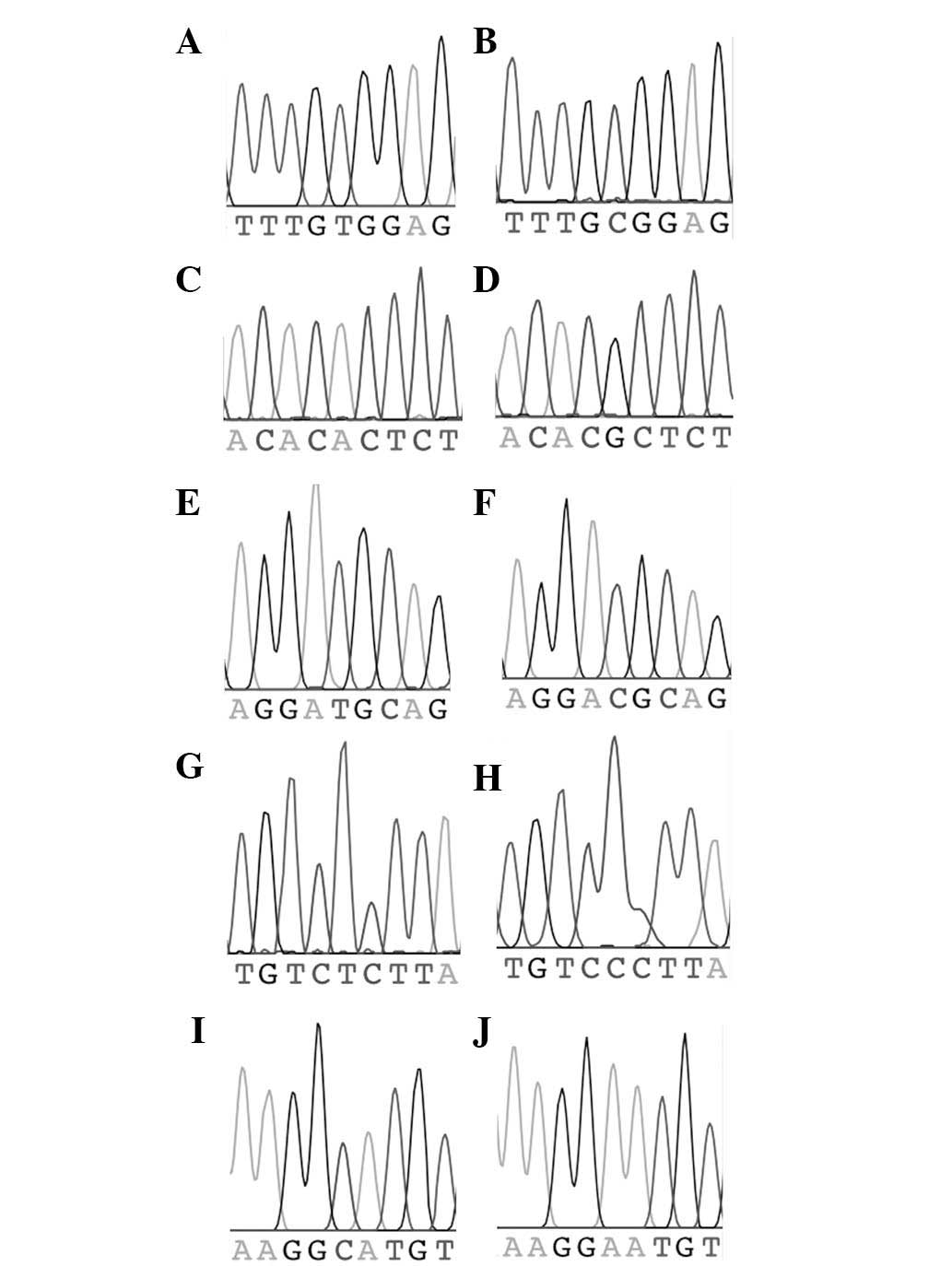
marker (two chromatograms per marker) are shown in Fig. 2.
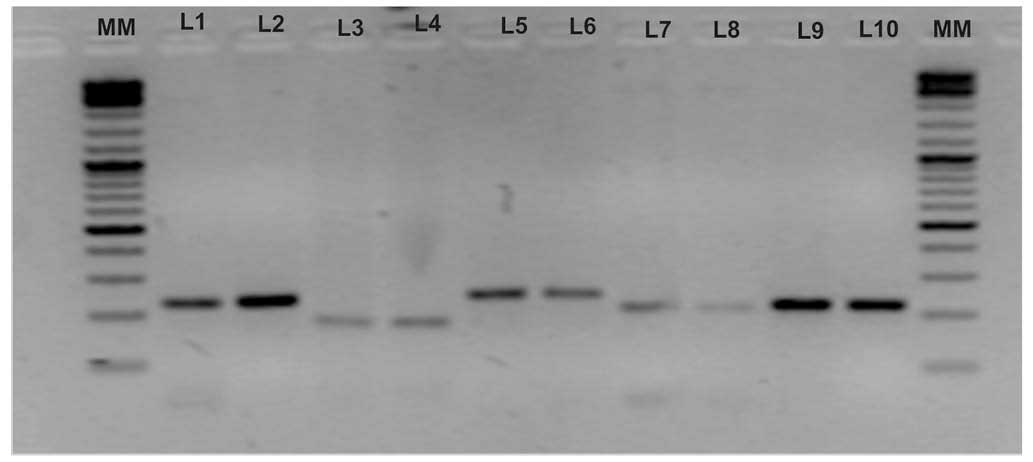 | Figure 1Amplification of allelic variants of
five genetic markers and their electrophoresis on a 2% agarose gel.
Lanes were loaded as follows: MM (molecular marker; Thermo Fisher
Scientific™ O'GeneRuler™ 100 bp Plus DNA Ladder); L1, variant T of
rs2981582 (223 bp); L2, variant C of rs2981582 (223 bp); L3,
variant A of rs1219648 (177 bp); L4, variant G of rs1219648 (177
bp); L5, variant T of rs8051542 (242 bp); L6, variant C of
rs8051542 (242 bp); L7, variant T of rs3803662 (210 bp); L8,
variant C of rs3803662 (210 bp); L9, variant C of rs889312 (219
bp); L10, variant A of rs889312 (219 bp). |
The baseline characteristics of 100 female patients
with BC are shown in Table II.
The mean age of all the patients at diagnosis was 46.1±11.63 years
(range, 20–67 years). Out of the total of 100 patients with BC,
only 24 cases were familial, and the remaining 76 cases were
sporadic, according to the medical history written in the patients'
files. Consanguineous marriages accounted for ~50% of the patients
(n=48). Overall, the most common UICC cancer stages (36) identified in the randomly selected
group of patients were stages 2 to 3, which accounted for 69% (30
and 39% respectively) of the cases, whereas stages 1 and 4
accounted for only 6 and 11%, respectively. The stage of the
remaining 14% of the cases was unknown. No patient presented at
stage 0 (carcinoma in situ). The BC patient samples
comprised 50 (50%) ER-positive tumors and 33 (33%) ER-negative
tumors, whereas the status of the remaining 17 (17%) was unknown;
39% of the patients with BC had PR-positive tumors and 46% had
PR-negative tumors, with 15% of the patients unknown; 37% of the
patients with BC had HER2-positive tumors and 26% had HER2-negative
tumors, with 10% of the patients untested. Triple-negative (i.e.
ER−, PR− and HER2−) patients with BC accounted for 27%, whereas 11%
of the patients had luminal A tumors (ER or PR+, HER2−) in randomly
collected blood samples.
 | Table IIBaseline characteristics of selected
variables in female breast cancer patients of Pakistan (n=100). |
Table II
Baseline characteristics of selected
variables in female breast cancer patients of Pakistan (n=100).
| Variable | Value |
|---|
| Age, years (mean ±
SD) | 46.1±11.63 (range
20–67) |
| History | |
| Sporadic | 76 |
| Familial | 24 |
| Consanguineous
marriages (out of total samples) | 48 |
| Clinical staging of
cancer (UICC)a (n=100) |
| Stage 0 (in
situ) | 0 (0.0%) |
| Stage 1 | 6 (6.00%) |
| Stage 2 | 30 (30.0%) |
| Stage 3 | 39 (39.0%) |
| Stage 4 | 11 (11.0%) |
| Unknown | 14 (14.0%) |
| Receptor
status |
| Estrogen receptor
(n=100) | |
| Positive | 50 (50%) |
| Negative | 33 (33%) |
| Unknown | 17 (17%) |
| Progesterone
receptor (n=100) | |
| Positive | 39 (39%) |
| Negative | 46 (46%) |
| Unknown | 15 (15%) |
| Human epidermal
growth factor receptor 2 (n=100) |
| Positive | 37 (37%) |
| Negative | 26 (26%) |
| Unknown | 02 (02%) |
|
Triple-negativeb | 27 (27%) |
| Luminal Ac | 11 (11%) |
Subsequently, two FGFR2 SNPs (rs1219648, rs2981582),
two TNRC9 SNPs (rs8051542, rs3803662) and one MAP3K1 SNP (rs889312)
were genotyped in the cases of Pakistani women with BC (90-100), as
well as control subjects. The Hardy-Weinberg equilibrium was
assessed for all five SNP genotypes in patients with BC and in
controls using SHEsis online software (35). The association of each genetic
marker with BC risk (P-values), as well as the total number of
samples analyzed in both case and control subjects, is shown in
Table III. Significant
associations were observed between two SNPs of FGFR2 [rs2981582
(P=0.005), rs1219648 (P=9.08e–006)], one SNP of TNRC9 [rs3803662
(P= 0.012)] and the BC risk, although no significant associations
were identified in the second SNP of TNRC9 (rs8051542) and the SNP
of MAP3K1 (rs889312). The most significant association with the BC
risk was observed for rs1219648 in the FGFR2 gene
(P=9.08e–006).
 | Table IIIThe association of FGFR2 (rs2981582
and rs1219648) and TNRC9 (rs3803662) with breast cancer risk in
Pakistani female patients. |
Table III
The association of FGFR2 (rs2981582
and rs1219648) and TNRC9 (rs3803662) with breast cancer risk in
Pakistani female patients.
| Gene | Marker | Case | Control | Fisher's
P-value |
|---|
| FGFR2 | rs2981582a | 100 | 100 | 0.005 |
| rs1219648a | 90 | 90 | 9.08e–006 |
| TNRC9 | rs8051542 | 96 | 90 | 0.506 |
| rs3803662a | 96 | 90 | 0.012 |
| MAP3K1 | rs889312 | 100 | 100 | 0.245 |
The distribution of SNP genotypes between the cancer
group and the healthy controls was compared using the Chi-squared
test to see which genotype of a particular marker is associated
with BC risk (Table IV). The
homozygote GG (P=0.000087), as well as heterozygote AG
(P=0.000018), genotypes of SNP rs1219648 in the FGFR2 gene
exhibited significant association with the risk of BC. However,
only homozygote CT genotype (P=0.001) of SNP rs2981582 in the FGFR2
gene was significantly associated with BC risk, and homozygote TT
genotype (P=0.097) did not show any association, as the number of
samples of this particular genotype was equal in both the case
(4) and control (4) groups. Similarly, only heterozygote CT
genotype (P=0.006) of SNP rs3803662 in the TNRC9 gene was
significantly associated with BC risk, and homozygote TT genotype
(P=0.378) did not reveal any association, as the number of samples
for this TT genotype was very low in the control group (1) and no samples were identified with
this genotype in the case group. No significant correlation was
identified between MAP3K1 SNP (rs889312) and BC risk on an
examination of its homozygote or heterozygote genotypes (Table IV).
 | Table IVAssociation of FGFR2, TNRC9 and
MAP3K1 genotypes with breast cancer in Pakistani women. |
Table IV
Association of FGFR2, TNRC9 and
MAP3K1 genotypes with breast cancer in Pakistani women.
| SNP ID | Genotype | Number
| OR (95% CI)b | P-valuec |
|---|
| Control | Case |
|---|
| rs2981582 | CC | 20 | 5 | Reference | – |
| CT | 76 | 91 | 1.197
(1.070–1.340) | 0.001a |
| TT | 4 | 4 | 2.667
(0.840–8.463) | 0.097 |
| rs1219648 | AA | 23 | 1 | Reference | – |
| AG | 57 | 76 | 1.385
(1.202–1.596) | 0.000018a |
| GG | 10 | 13 | 3.064
(1.790–5.245) | 0.000087a |
| rs8051542 | CC | 35 | 45 | Reference | – |
| CT | 38 | 37 | 0.867
(0.626–1.199) | 0.389 |
| TT | 17 | 14 | 0.726
(0.398–1.324) | 0.294 |
| rs3803662 | CC | 64 | 50 | Reference | – |
| CT | 25 | 46 | 1.706
(1.152–2.526) | 0.006a |
| TT | 1 | 0 | – | 0.378 |
| rs889312 | AA | 41 | 40 | Reference | – |
| CA | 44 | 52 | 1.092
(0.831–1.434) | 0.526 |
| CC | 15 | 8 | 0.622
(0.289–1.339) | 0.215 |
Furthermore, genotypes were sorted on the basis of
various clinicopathological characteristics of BC for each marker,
and the association of the genetic marker was assessed in
stratified groups. The genetic variants exhibiting statistically
significant differences with respect to associations of various
clinicopathological characteristics with BC risk are shown in
Table V. Analyses stratified by ER
status revealed that SNP rs1219648 of the FGFR2 gene remained
significantly associated with BC risk in ER-positive (P=0.042) and
ER-negative (P=0.003) tumors. However, neither SNP rs2981582 of the
FGFR2 gene nor SNP rs3803662 of the TNRC9 gene revealed significant
associations with either ER-positive or ER-negative tumors. On
considering the PR status, SNP rs1219648 of the FGFR2 gene again
remained significantly associated with BC risk in PR-positive
(P=0.090) and PR-negative (P=0.0007) tumors. However, neither SNP
rs2981582 of the FGFR2 gene nor SNP rs3803662 of the TNRC9 gene
revealed a significant association with either PR-positive or
PR-negative tumors. In the case of HER2 carriers, SNP rs1219648 of
FGFR2 exhibited a significant association with a higher BC risk in
HER2-positive (P=0.0009) tumors. However, SNP rs3803662 of the
TNRC9 gene revealed differing levels of association, and it was
only associated significantly with BC risk in HER2-negative
patients (P=0.023). By considering triple-negative cases
(ER-negative, PR-negative and HER2-negative), SNP rs1219648 of
FGFR2 (P=0.004) and rs3803662 of TNRC9 (P=0.014) revealed a
significant association with BC risk, although no significant
association was observed in the case of SNP rs2981582 of FGFR2 gene
(P=0.058).
 | Table VThe association of FGFR2 and TNRC9
variants with breast cancer risk considering various
clinicopathological characteristics. |
Table V
The association of FGFR2 and TNRC9
variants with breast cancer risk considering various
clinicopathological characteristics.
A, ER+/−
|
|---|
| SNP | Controls (n) | ER-positive cases
| ER-negative cases
|
|---|
| Cases (n) | P-value | OR (95% CI) | Cases (n) | P-value | OR (95% CI) |
|---|
| rs3803662 | 72 | 37 | 0.139 | 0.604
(0.298–1.225) | 13 | 0.170 | 0.489
(0.184–1.301) |
| rs2981582 | 66 | 33 | 0.111 | 0.709
(0.360–1.393) | 08 | 0.290 | 0.650
(0.229–1.839) |
| rs1219648 | 32 | 31 | 0.042a | 1.138
(0.539–2.404) | 16 | 0.003a | 1.476
(0.607–3.584) |
B, PR+/−
|
|---|
| SNP | Controls (n) | PR-positive cases
| PR-negative cases
|
|---|
| Cases (n) | P-value | OR (95% CI) | Cases (n) | P-value | OR (95% CI) |
|---|
| rs3803662 | 72 | 31 | 0.196 | 0.618
(0.292–1.307) | 20 | 0.069 | 0.475
(0.207–1.089) |
| rs2981582 | 66 | 20 | 0.165 | 0.718
(0.352–1.464) | 11 | 0.185 | 0.650
(0.262–1.607) |
| rs1219648 | 32 | 27 | 0.090a | 1.095
(0.505–2.372) | 21 | 0.0007a | 1.476
(0.650–3.352) |
C, HER2+/−
|
|---|
| SNP | Controls (n) | HER2-positive cases
| HER2-negative cases
|
|---|
| Cases (n) | P-value | OR (95% CI) | Cases (n) | P-value | OR (95% CI) |
|---|
| rs380366272 | 72 | 33 | 0.410 | 0.735
(0.344–1.568) | 12 | 0.023a | 0.360
(0.137–0.944) |
| rs2981582 | 66 | 20 | 0.051 | 0.650
(0.319–1.323) | 08 | 0.290 | 0.650
(0.229–1.839) |
| rs1219648 | 32 | 31 | 0.0009a | 1.476
(0.701–3.108) | 12 | 0.226 | 0.885
(0.327–2.395) |
D, sporadic and
familial
|
|---|
| SNP | Controls (n) | Sporadic cases
| Familial cases
|
|---|
| Cases (n) | P-value | OR (95% CI) | Cases (n) | P-value | OR (95% CI) |
|---|
| rs3803662 | 72 | 76 | 0.035a | 0.582
(0.322–1.051) | 21 | 0.101 | 0.508
(0.222–1.158) |
| rs2981582 | 66 | 57 | 0.002a | 0.650
(0.391–1.078) | 10 | 0.215 | 0.650
(0.253–1.669) |
| rs1219648 | 32 | 66 | 0.002a | 1.181
(0.644–2.164) | 21 | 0.008a | 1.328
(0.606–2.911) |
E, triple-negative
|
|---|
| SNP | Controls (n) | Triple-negative
cases
|
|---|
| Cases (n) | P-value | OR (95% CI) |
|---|
| rs3803662 | 72 | 25 | 0.014a | 0.420
(0.197–0.896) |
| rs2981582 | 66 | 19 | 0.058 | 0.650
(0.314–1.342) |
| rs1219648 | 32 | 24 | 0.004a | 1.249
(0.565–2.761) |
The sporadic and familial breast cases were sorted
to assess whether the association of markers with BC risk would be
significant, based on the family history of the patients. SNP
rs1219648 of the FGFR2 gene again remained significantly associated
with BC risk in sporadic (P=0.002) and familial (P=0.008) cases,
although the other two SNPs (rs2981582 of FGFR2 and rs3803662 of
TNRC9) exhibited significant associations with BC risk only in
sporadic cases (P=0.002, P=0.035 respectively).
Another important issue taken into consideration
during the study was to assess whether women carrying risk alleles
at both loci of the identical marker are at an even greater risk of
BC compared with those carrying only one risk allele, and whether
any effects would ensue as a consequence of their epistatic
interactions. Therefore, combined effects (haplotypes) of the two
SNPs of the FGFR2 gene (rs1219648, rs2981582), as well as the two
SNPs of the TNRC9 gene (rs8051542, rs3803662), were analyzed
(Table VI). The analysis
identified four common haplotypes in the SNPs of FGFR2, with
frequencies >0.03, and all haplotypes were significantly
associated with BC risk, providing evidence that women carrying
more alleles associated with risk have a greater chance of
developing BC. However, in the case of haplotyping of TNRC9 SNPs,
of the four identified haplotypes, the CT haplotype (OR, 3.519; 95%
CI, 1.769–6.999; P=0.00017) revealed the most significant
association with risk of BC.
 | Table VIThe association of FGFR2 and TNRC9
haplotypes with breast cancer risk. |
Table VI
The association of FGFR2 and TNRC9
haplotypes with breast cancer risk.
| Gene | Marker | Haplotypea | Case frequency | Control
frequency | P-value | OR (95% CI) |
|---|
| FGFR2 | rs2981582 | C Ab | 6.07 (0.034) | 82.56 (0.459) | 3.22e–015 | 0.041
(0.017–0.097) |
| C Gb | 85.93 (0.477) | 22.44 (0.125) | 3.46e–013 | 6.414
(3.775–10.900) |
| rs1219648 | T Ab | 71.93 (0.400) | 20.44 (0.114) | 5.60e–010 | 5.196
(3.002–8.994) |
| T Gb | 16.07 (0.089) | 54.56 (0.303) | 3.38e–007 | 0.225
(0.123–0.412) |
| TNRC9 | rs8051542 | C C | 88.86 (0.463) | 96.15 (0.534) | 0.168778 | 0.751
(0.500–1.129) |
| C Tb | 38.14 (0.199) | 11.85 (0.066) | 0.000176 | 3.519
(1.769–6.999) |
| rs3803662 | T C | 57.14 (0.298) | 56.85 (0.316) | 0.703778 | 0.918
(0.591–1.427) |
| T T | 7.86 (0.041) | 15.15 (0.084) | 0.083435 | 0.464
(0.191–1.127) |
Discussion
In this case-control study of BC in Pakistani women,
significant associations were observed between two SNPs of FGFR2
[rs1219648: P=9.08e-006 (GG, P=0.000087, AG, P=0.000018) and
rs2981582: P=0.005 (CT, P=0.001, TT, P=0.097)]. These results are
consistent with results obtained from other Asian regions,
including China (16,32,37),
Japan (38) and India (18). Furthermore, the SNPs rs1219648 and
rs2981582 of FGFR2 have been consistently associated with BC risk
in several other ethnic groups, including European (39), Hispanic and non-Hispanic Caucasian
women from Southwestern United States (40), African American (29,41)
and Tunisian (42) women
populations. The two SNPs (rs1219648 and rs2981582) belong to
intronic region (intron 2) of the FGFR2 gene, and the precise
mechanism that would explain how FGFR2 risk alleles induce
upregulation of the expression of FGFR2 has yet to be fully
elucidated (32). However, it has
been reported in a couple of previous studies that these variants
upregulate the expression of FGFR2 in BC tissues by acting as
enhancer regions, which may result in tumor formation (18,43).
The aberrant expression of nine different isoforms of FGFR2 as a
result of alternative splicing has shown the activation of signal
transduction and development of BC (14,44).
In addition, a high degree of conservation in intron 2 of FGFR2 in
mammals, and the presence of several putative transcription-factor
binding sites (15) in the
proximate regions of the significant SNPs, suggest that these SNPs
may exert a significant role in tumor development (18).
The additional classifications of BC based on
clinicopathological characteristics, including ER, PR and HER2
status, have been utilized to understand the etiology of the
heterogeneous tumor, which may be helpful in terms of elucidating
the mechanisms of carcinogenesis and in improving the prevention
and treatment of cancer (45).
Therefore, the associations between the two variants of FGFR2 and
clinicopathological characteristics of BC were further assessed.
Analyses stratified by the status of the ER and PR in the present
study revealed that SNP rs1219648 of the FGFR2 gene remained
significantly associated with BC risk in the two intrinsic
subtypes, including ER-positive (P=0.042) and ER-negative (P=0.003)
tumors, as well as in PR-positive (P=0.090) and PR negative
(P=0.0007) tumors, consistent with the results of certain previous
studies (16,18,32,37).
The second SNP of the FGFR2 gene, rs2981582, did not show
significant associations with either ER- and PR-positive or ER- and
PR-negative tumors, although SNP rs2981582 was significantly
associated in the whole sample set (non-stratified) of cases. The
lack of any association may be the result of a low number of
samples in each intrinsic subtype following stratification,
considering ER, PR and HER2.
In the case of TNRC9 (TOX3) gene variants, a
significant association was observed between SNP rs3803662 and the
risk of BC (P=0.012), although no associations were identified in
SNP rs8051542 of TNRC9 in Pakistani women, consistent with the
results of a meta-analysis study (46). Previously published reports have
revealed a positive correlation of SNP rs3803662 of TNRC9 with BC
risk in different ethnic groups (10,15).
However, repeated studies of the identical SNP among European,
African-American and East Asian populations reported contradictory
results (23–25,28).
Furthermore, the role of these SNPs as BC susceptibility variants
in TNRC9 has yet to be determined. However, the overexpression of
TNRC9 in BC, particularly in advanced BC, has been reported in a
few studies (47,48), and its amplification has been
associated with reduced disease-free and metastasis-free survival
rates. The inverse correlation of the expression of TNRC9 and BRCA1
provided further evidence for the involvement of TNRC9 in the
development of BC (19). Analyses
stratified by ER and PR status revealed that SNP rs3803662 of the
TNRC9 gene did not exhibit significant associations with any
intrinsic subtype (ER- and PR-positive, as well as ER- and
PR-negative tumors) in the present study. Similar observations of
the genetic variant rs3803662 in TNRC9 have been reported in a
previous study on a Chinese population (49). However, a study that included
12,974 ER-positive and 3,765 ER-negative cases reported the
association of rs3803662 with BC risk in the two tumor subtypes
(ER+ and ER− tumors), in contrast with our results (50). These controversies in the
literature regarding the association of TNRC9 SNPs with risk of BC,
as well as with intrinsic subtypes in different ethnic groups,
exist due to the following two major reasons: First, genetic
factors differ according to ethnicity, and secondly, larger sample
sizes are required to assess gene-gene and gene-environment
interactions to signify a powerful BC risk in the population
(49).
Regarding the variant of MAP3K1, no significant
association of SNP rs889312 (P=0.245) with the risk of BC in
Pakistani women was observed in the present study. This finding is
also relevant to previous studies for women of African ancestry
(29–31), demonstrating a poor association of
the SNP with BC. However, the rs889312-C allele of MAP3K1 has been
reported as a risk factor for the development of BC in European and
Asian ancestry populations (27),
contrary to the results in the present study.
All three BC-associated genetic variants in the
whole sample set (rs2981582, rs1219648, rs3803662) also remained
associated significantly with an increased risk of BC in the
sporadic group of patients when sorted on the basis of the family
history of BC in our population. This finding regarding the
involvement of these genetic variants for sporadic BC in our
population is consistent with previous observations made in Chinese
women (37), in Chinese Han Women
(32) and in sporadic
post-menopausal women of European ancestry with respect to the SNPs
of FGFR2 (rs2981582, rs1219648) (51). Furthermore, the overexpression of
the FGFR2 and TNRC9 genes in sporadic patients with BC compared
with controls further supports the greater involvement of these
susceptibility loci in a predisposition to sporadic BC (19).
In conclusion, the present study has provided
evidence revealing a significant association of FGFR2 intron 2 SNPs
(rs2981582 and rs1219648) and TNRC9 SNP (rs3803662) with BC among
Pakistani women. Along with variable interactions of these SNPs
with different clinicopathological characteristics, all three
genetic variants (rs2981582, rs1219648, rs3803662) revealed a
significant association with increased risk of sporadic BC in this
population. In addition, there was an increased effect (stronger
significant association) between haplotype combinations of the two
SNPs of FGFR2 (rs2981582 and rs1219648) with BC risk in Pakistani
women. Further studies of larger data sets, along with
subcategorization by clinical parameters, are required to confirm
the role of these variants in intrinsic subtypes of BC in Pakistan
that may help to improve our understanding of the genetic
heterogeneity in this complex disease in our population.
Abbreviations:
|
BC
|
breast cancer
|
|
BRCA1/2
|
breast cancer 1/2
|
|
SNP
|
single nucleotide polymorphism
|
|
FGFR2
|
fibroblast growth factor receptor
2
|
|
GWASs
|
genome-wide association studies
|
|
TNRC9
|
trinucleotide-repeat-containing 9
|
|
MAP3K1
|
mitogen-activated protein kinase
kinase kinase 1; ER
|
|
PR
|
progesterone receptor
|
|
HER2
|
human epidermal growth factor receptor
2
|
|
PCR
|
polymerase chain reaction
|
|
ORs
|
odds ratios
|
|
CIs
|
confidence intervals
|
Acknowledgments
The authors would like to thank the women, whether
patients or healthy control subjects, who were involved in this
study, and acknowledge the efforts and contribution of all the
support staff (Bioscience Laboratory, CIIT, Sahiwal, School of
Biological Sciences, University of the Punjab, Lahore and INMOL
hospital) who made the study possible. We also thank Cancer
Research Group Foundation, Lahore for providing funding.
References
|
1
|
Parkin DM, Bray F, Ferlay J and Pisani P:
Global cancer statistics, 2002. CA Cancer J Clin. 55:74–108. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hortobagyi GN, de la Garza Salazar J,
Pritchard K, Amadori D, Haidinger R, Hudis CA, Khaled H, Liu MC,
Martin M, Namer M, et al: The global breast cancer burden:
Variations in epidemiology and survival. Clin Breast Cancer.
6:391–401. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Shaukat U, Toor M, Ahmad B, Fazal S and
Mehmood N: Genetic and computational analysis of Tgfb1 & Fgfr2
polymorphism in correlation to breast cancer susceptibility in
Pakistani women. J Cancer Sci Ther. 6:433–439. 2014.
|
|
4
|
Lichtenstein P, Holm NV, Verkasalo PK,
Iliadou A, Kaprio J, Koskenvuo M, Pukkala E, Skytthe A and Hemminki
K: Environmental and heritable factors in the causation of
cancer-analyses of cohorts of twins from Sweden, Denmark and
Finland. N Engl J Med. 343:78–85. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Thompson D and Easton D: The genetic
epidemiology of breast cancer genes. J Mammary Gland Biol
Neoplasia. 9:221–236. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Antoniou AC and Easton DF: Models of
genetic susceptibility to breast cancer. Oncogene. 25:5898–5905.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Pharoah PD, Antoniou A, Bobrow M, Zimmern
RL, Easton DF and Ponder BA: Polygenic susceptibility to breast
cancer and implications for prevention. Nat Genet. 31:33–36. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Han W, Woo JH, Yu JH, Lee MJ, Moon HG,
Kang D and Noh DY: Common genetic variants associated with breast
cancer in Korean women and differential susceptibility according to
intrinsic subtype. Cancer Epidemiol Biomarkers Prev. 20:793–798.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hindorff LA, Sethupathy P, Junkins HA,
Ramos EM, Mehta JP, Collins FS and Manolio TA: Potential etiologic
and functional implications of genome-wide association loci for
human diseases and traits. Proc Natl Acad Sci USA. 106:9362–9367.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Fanale D, Amodeo V, Corsini LR, Rizzo S,
Bazan V and Russo A: Breast cancer genome-wide association studies:
There is strength in numbers. Oncogene. 31:2121–2128. 2012.
View Article : Google Scholar
|
|
11
|
Nordgard SH, Johansen FE, Alnaes GI, Naume
B, Børresen-Dale AL and Kristensen VN: Genes harbouring
susceptibility SNPs are differentially expressed in the breast
cancer subtypes. Breast Cancer Res. 9:1132007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Easton DF and Eeles RA: Genome-wide
association studies in cancer. Hum Mol Genet. 17:R109–R115. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Grose R and Dickson C: Fibroblast growth
factor signaling in tumorigenesis. Cytokine Growth Factor Rev.
16:179–186. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Moffa AB and Ethier SP: Differential
signal transduction of alternatively spliced FGFR2 variants
expressed in human mammary epithelial cells. J Cell Physiol.
210:720–731. 2007. View Article : Google Scholar
|
|
15
|
Easton DF, Pooley KA, Dunning AM, Pharoah
PD, Thompson D, Ballinger DG, Struewing JP, Morrison J, Field H,
Luben R, et al: Genome-wide association study identifies novel
breast cancer susceptibility loci. Nature. 447:1087–1093. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Liu CL, Hu XP, Guo WD, Yang L, Dang J and
Jiao HY: Case-control study on the fibroblast growth factor
receptor 2 gene polymorphisms associated with breast cancer in
Chinese han women. J Breast Cancer. 16:366–371. 2013. View Article : Google Scholar
|
|
17
|
Saadatian Z, Gharesouran J, Ghojazadeh M,
Ghohari-Lasaki S, Tarkesh-Esfahani N and Mohaddes Ardebili SM:
Association of rs1219648 in FGFR2 and rs1042522 in TP53 with
premenopausal breast cancer in an Iranian Azeri population. Asian
Pac J Cancer Prev. 15:7955–7958. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Siddiqui S, Chattopadhyay S, Akhtar MS,
Najm MZ, Deo SV, Shukla NK and Husain SA: A study on genetic
variants of fibroblast growth factor receptor 2 (FGFR2) and the
risk of breast cancer from North India. PLoS One. 9:e1104262014.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Shan J, Dsouza SP, Bakhru S, Al-Azwani EK,
Ascierto ML, Sastry KS, Bedri S, Kizhakayil D, Aigha II, Malek J,
et al: TNRC9 downregulates BRCA1 expression and promotes breast
cancer aggressiveness. Cancer Res. 73:2840–2849. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Mcinerney N, Colleran G, Rowan A, Walther
A, Barclay E, Spain S, Jones AM, Tuohy S, Curran C, Miller N, et
al: Low penetrance breast cancer predisposition SNPs are site
specific. Breast Cancer Res Treat. 117:151–159. 2009. View Article : Google Scholar
|
|
21
|
Chalabi N, Bernard-Gallon DJ, Bignon YJ;
Breast Med Consortium; Kwiatkowski F, Agier M, Vidal V,
Laplace-Chabaud V, Sylvain-Vidal V, Bertholet V, et al: Comparative
clinical and transcriptomal profiles of breast cancer between
French and South Mediterranean patients show minor but
significative biological differences. Cancer Genomics Proteomics.
5:253–261. 2008.
|
|
22
|
Huijts PE, Vreeswijk MP, Kroeze-Jansema
KH, Jacobi CE, Seynaeve C, Krol-Warmerdam EM, Wijers-Koster PM,
Blom JC, Pooley KA, Klijn JG, et al: Clinical correlates of
low-risk variants in FGFR2, TNRC9, MAP3K1, LSP1 and 8q24 in a Dutch
cohort of incident breast cancer cases. Breast Cancer Res.
9:R782007. View
Article : Google Scholar : PubMed/NCBI
|
|
23
|
Sueta A, Ito H, Kawase T, Hirose K, Hosono
S, Yatabe Y, Tajima K, Tanaka H, Iwata H, Iwase H and Matsuo K: A
genetic risk predictor for breast cancer using a combination of
low-penetrance polymorphisms in a Japanese population. Breast
Cancer Res Treat. 132:711–721. 2012. View Article : Google Scholar
|
|
24
|
Ruiz-Narváez EA, Rosenberg L, Cozier YC,
Cupples LA, Adams-Campbell LL and Palmer JR: Polymorphisms in the
TOX3/LOC643714 locus and risk of breast cancer in African-American
women. Cancer Epidemiol Biomarkers Prev. 19:1320–1327. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Long J, Shu XO, Cai Q, Gao YT, Zheng Y, Li
G, Li C, Gu K, Wen W, Xiang YB, et al: Evaluation of breast cancer
susceptibility loci in Chinese women. Cancer Epidemiol Biomarkers
Prev. 19:2357–2365. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Pham TT, Angus SP and Johnson GL: MAP3K1:
Genomic alterations in cancer and function in promoting cell
survival or apoptosis. Genes Cancer. 4:419–426. 2013. View Article : Google Scholar
|
|
27
|
Hirschhorn JN, Lohmueller K, Byrne E and
Hirschhorn K: A comprehensive review of genetic association
studies. Genet Med. 4:45–61. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kim HC, Lee JY, Sung H, Choi JY, Park SK,
Lee KM, Kim YJ, Go MJ, Li L, Cho YS, et al: A genome-wide
association study identifies a breast cancer risk variant in ERBB4
at 2q34: Results from the Seoul breast cancer study. Breast Cancer
Res. 14:R562012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Barnholtz-Sloan JS, Shetty PB, Guan X,
Nyante SJ, Luo J, Brennan DJ and Millikan RC: FGFR2 and other loci
identified in genome-wide association studies are associated with
breast cancer in African-American and younger women.
Carcinogenesis. 31:1417–1423. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zheng Y, Ogundiran TO, Falusi AG,
Nathanson KL, John EM, Hennis AJ, Ambs S, Domchek SM, Rebbeck TR,
Simon MS, et al: Fine mapping of breast cancer genome-wide
association studies loci in women of African ancestry identifies
novel susceptibility markers. Carcinogenesis. 34:1520–1528. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Chen F, Chen GK, Millikan RC, John EM,
Ambrosone CB, Bernstein L, Zheng W, Hu JJ, Ziegler RG, Deming SL,
et al: Fine-mapping of breast cancer susceptibility loci
characterizes genetic risk in African Americans. Hum Mol Genet.
20:4491–4503. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Fu F, Wang C, Huang M, Song C, Lin S and
Huang H: Polymorphisms in second intron of the FGFR2 gene are
associated with the risk of early-onset breast cancer in Chinese
Han women. Tohoku J Exp Med. 226:221–229. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Maniatis T, Fritsch EF and Sambrook J:
1982 Molecular cloning: A laboratory manual. Cold Spring Harbor
Laboratory; Cold Spring Harbor, New York:
|
|
34
|
Hirotsu N, Murakami N, Kashiwagi T, Ujiie
K and Ishimaru K: Protocol: A simple gel-free method for SNP
genotyping using allele-specific primers in rice and other plant
species. Plant Methods. 6:122010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Li Z, Zhang Z, He Z, Tang W, Li T, Zeng Z,
He L and Shi Y: A partition-ligation-combination-subdivision EM
algorithm for haplotype inference with multiallelic markers: Update
of the SHEsis (http://analysis.bio-x.cn).
Cell Res. 19:519–523. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Haffty BG, Buchholz TA and Perez CA: Early
stage breast cancer. Principles and Practice of Radiation Oncology.
Halperin EC, Perez CA and Brady LW: 5th ed. J. B. Lippincott &
Co.; Philadelphia: pp. 1175–1291. 2008
|
|
37
|
Liang J, Chen P, Hu Z, Zhou X, Chen L, Li
M, Wang Y, Tang J, Wang H and Shen H: Genetic variants in
fibroblast growth factor receptor 2 (FGFR2) contribute to
susceptibility of breast cancer in Chinese women. Carcinogenesis.
29:2341–2346. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kawase T, Matsuo K, Suzuki T, Hiraki A,
Watanabe M, Iwata H, Tanaka H and Tajima K: FGFR2 intronic
polymorphisms interact with reproductive risk factors of breast
cancer: Results of a case control study in Japan. Int J Cancer.
125:1946–1952. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Stacey SN, Manolescu A, Sulem P,
Thorlacius S, Gudjonsson SA, Jonsson GF, Jakobsdottir M,
Bergthorsson JT, Gudmundsson J, Aben KK, et al: Common variants on
chromosome 5p12 confer susceptibility to estrogen receptor-positive
breast cancer. Nat Genet. 40:703–706. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Slattery ML, Baumgartner KB, Giuliano AR,
Byers T, Herrick JS and Wolff RK: Replication of five
GWAS-identified loci and breast cancer risk among Hispanic and
non-Hispanic white women living in the Southwestern United States.
Breast Cancer Res Treat. 129:531–539. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zheng W, Cai Q, Signorello LB, Long J,
Hargreaves MK, Deming SL, Li G, Li C, Cui Y and Blot WJ: Evaluation
of 11 breast cancer susceptibility loci in African-American women.
Cancer Epidemiol Biomarkers Prev. 18:2761–2764. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Shan J, Mahfoudh W, Dsouza SP, Hassen E,
Bouaouina N, Abdelhak S, Benhadjayed A, Memmi H, Mathew RA, Aigha
II, et al: Genome-wide association studies (GWAS) breast cancer
susceptibility loci in Arabs: Susceptibility and prognostic
implications in Tunisians. Breast Cancer Res Treat. 135:715–724.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Huijts PE, van Dongen M, de Goeij MC, van
Moolenbroek AJ, Blanken F, Vreeswijk MP, de Kruijf EM, Mesker WE,
van Zwet EW, Tollenaar RA, et al: Allele-specific regulation of
FGFR2 expression is cell type-dependent and may increase breast
cancer risk through a paracrine stimulus involving FGF10. Breast
Cancer Res. 13:R722011. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Raskin L, Pinchev M, Arad C, Lejbkowicz F,
Tamir A, Rennert HS, Rennert G and Gruber SB: FGFR2 is a breast
cancer susceptibility gene in Jewish and Arab Israeli population.
Cancer Epidemiol Biomarkers Prev. 17:1060–1065. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Broeks A, Schmidt MK, Sherman ME, Couch
FJ, Hopper JL, Dite GS, Apicella C, Smith LD, Hammet F, Southey MC,
et al: Low penetrance breast cancer susceptibility loci are
associated with specific breast tumor subtypes: Findings from the
breast cancer association consortium. Hum Mol Genet. 20:3289–3303.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Chen MB, Wu XY, Shen W, Wei MX, Li C, Cai
B, Tao GQ and Lu PH: Association between polymorphisms of
trinucleotide repeat containing 9 gene and breast cancer risk:
Evidence from 62,005 subjects. Breast Cancer Res Treat.
126:177–183. 2011. View Article : Google Scholar
|
|
47
|
He X, Yao G, Li F, Li M and Yang X:
Risk-association of five SNPs in TOX3/LOC643714 with breast cancer
in southern China. Int J Mol Sci. 15:2130–2141. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Garcia-Closas M, Hall P, Nevanlinna H,
Pooley K, Morrison J, Richesson DA, Bojesen SE, Nordestgaard BG,
Axelsson CK, Arias JI, et al: Heterogeneity of breast cancer
associations with five susceptibility loci by clinical and
pathological characteristics. PLoS Genet. 4:e10000542008.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Hunter DJ, Kraft P, Jacobs KB, Cox DG,
Yeager M, Hankinson SE, Wacholder S, Wang Z, Welch R, Hutchinson A,
et al: A genome-wide association study identifies alleles in FGFR2
associated with risk of sporadic postmenopausal breast cancer. Nat
Genet. 39:870–874. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
50
|
Shan J, Dsouza SP, Bakhru S, Al-Azwani EK,
Ascierto ML, Sastry KS, Bedri S, Kizhakayil D, Aigha II, Malek J,
et al: TNRC9 downregulates BRCA1 expression and promotes breast
cancer aggressiveness. Cancer Res. 73:2840–2849. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Chalabi N, Bernard-Gallon DJ, Bignon YJ,
Kwiatkowski F, Agier M, Vidal V, Laplace-Chabaud V, Sylvain-Vidal
V, Bertholet V, De Longueville F, et al: Comparative clinical and
transcriptomal profiles of breast cancer between French and South
Mediterranean patients show minor but significative biological
differences. Cancer Genomics Proteomics. 5:253–261. 2008.
|