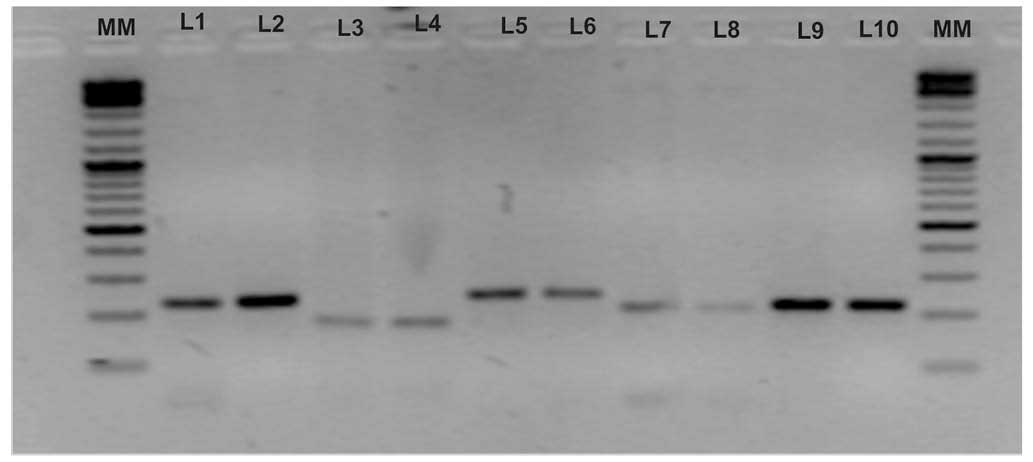
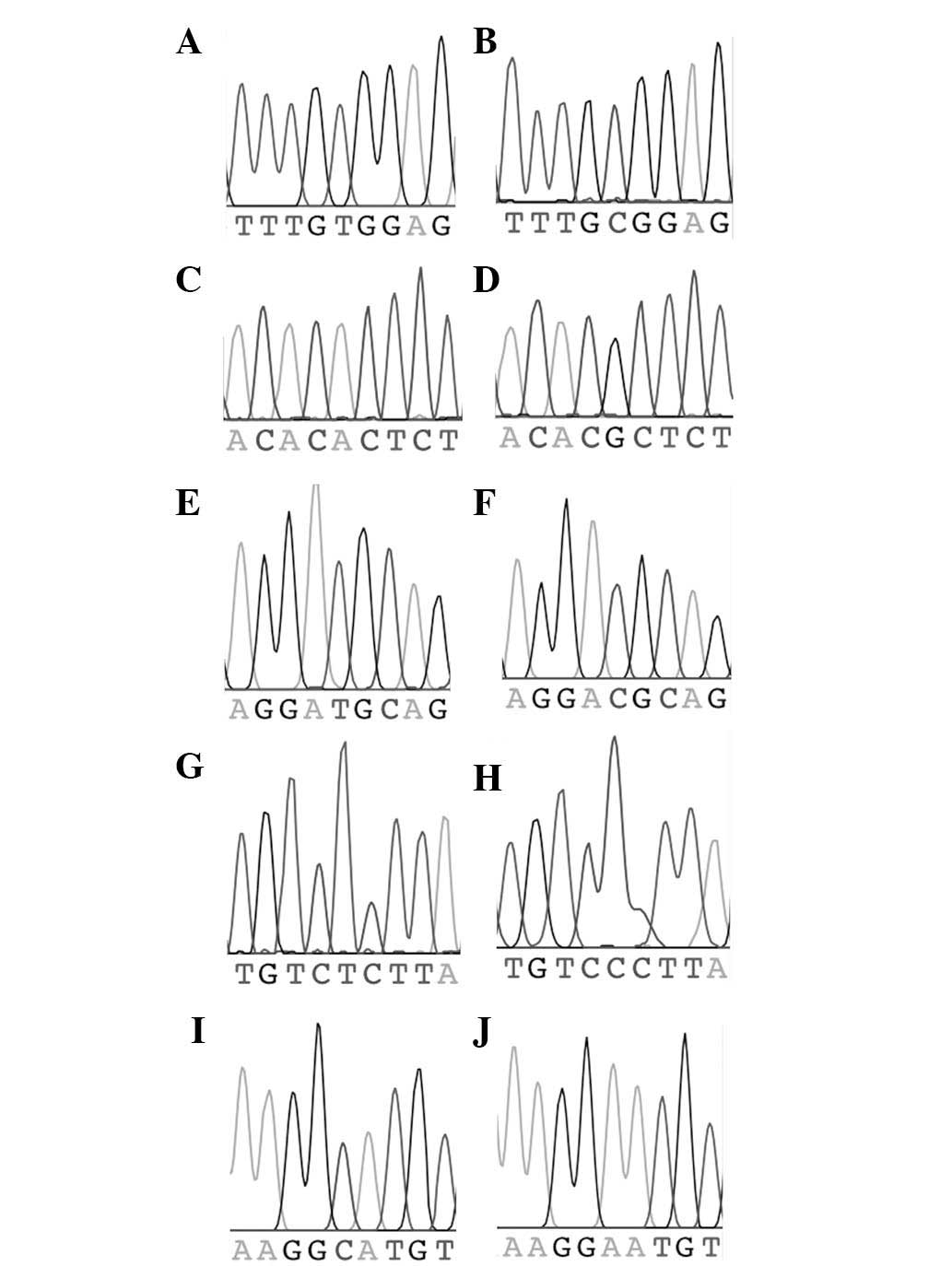
|
1
|
Parkin DM, Bray F, Ferlay J and Pisani P:
Global cancer statistics, 2002. CA Cancer J Clin. 55:74–108. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hortobagyi GN, de la Garza Salazar J,
Pritchard K, Amadori D, Haidinger R, Hudis CA, Khaled H, Liu MC,
Martin M, Namer M, et al: The global breast cancer burden:
Variations in epidemiology and survival. Clin Breast Cancer.
6:391–401. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Shaukat U, Toor M, Ahmad B, Fazal S and
Mehmood N: Genetic and computational analysis of Tgfb1 & Fgfr2
polymorphism in correlation to breast cancer susceptibility in
Pakistani women. J Cancer Sci Ther. 6:433–439. 2014.
|
|
4
|
Lichtenstein P, Holm NV, Verkasalo PK,
Iliadou A, Kaprio J, Koskenvuo M, Pukkala E, Skytthe A and Hemminki
K: Environmental and heritable factors in the causation of
cancer-analyses of cohorts of twins from Sweden, Denmark and
Finland. N Engl J Med. 343:78–85. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Thompson D and Easton D: The genetic
epidemiology of breast cancer genes. J Mammary Gland Biol
Neoplasia. 9:221–236. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Antoniou AC and Easton DF: Models of
genetic susceptibility to breast cancer. Oncogene. 25:5898–5905.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Pharoah PD, Antoniou A, Bobrow M, Zimmern
RL, Easton DF and Ponder BA: Polygenic susceptibility to breast
cancer and implications for prevention. Nat Genet. 31:33–36. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Han W, Woo JH, Yu JH, Lee MJ, Moon HG,
Kang D and Noh DY: Common genetic variants associated with breast
cancer in Korean women and differential susceptibility according to
intrinsic subtype. Cancer Epidemiol Biomarkers Prev. 20:793–798.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hindorff LA, Sethupathy P, Junkins HA,
Ramos EM, Mehta JP, Collins FS and Manolio TA: Potential etiologic
and functional implications of genome-wide association loci for
human diseases and traits. Proc Natl Acad Sci USA. 106:9362–9367.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Fanale D, Amodeo V, Corsini LR, Rizzo S,
Bazan V and Russo A: Breast cancer genome-wide association studies:
There is strength in numbers. Oncogene. 31:2121–2128. 2012.
View Article : Google Scholar
|
|
11
|
Nordgard SH, Johansen FE, Alnaes GI, Naume
B, Børresen-Dale AL and Kristensen VN: Genes harbouring
susceptibility SNPs are differentially expressed in the breast
cancer subtypes. Breast Cancer Res. 9:1132007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Easton DF and Eeles RA: Genome-wide
association studies in cancer. Hum Mol Genet. 17:R109–R115. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Grose R and Dickson C: Fibroblast growth
factor signaling in tumorigenesis. Cytokine Growth Factor Rev.
16:179–186. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Moffa AB and Ethier SP: Differential
signal transduction of alternatively spliced FGFR2 variants
expressed in human mammary epithelial cells. J Cell Physiol.
210:720–731. 2007. View Article : Google Scholar
|
|
15
|
Easton DF, Pooley KA, Dunning AM, Pharoah
PD, Thompson D, Ballinger DG, Struewing JP, Morrison J, Field H,
Luben R, et al: Genome-wide association study identifies novel
breast cancer susceptibility loci. Nature. 447:1087–1093. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Liu CL, Hu XP, Guo WD, Yang L, Dang J and
Jiao HY: Case-control study on the fibroblast growth factor
receptor 2 gene polymorphisms associated with breast cancer in
Chinese han women. J Breast Cancer. 16:366–371. 2013. View Article : Google Scholar
|
|
17
|
Saadatian Z, Gharesouran J, Ghojazadeh M,
Ghohari-Lasaki S, Tarkesh-Esfahani N and Mohaddes Ardebili SM:
Association of rs1219648 in FGFR2 and rs1042522 in TP53 with
premenopausal breast cancer in an Iranian Azeri population. Asian
Pac J Cancer Prev. 15:7955–7958. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Siddiqui S, Chattopadhyay S, Akhtar MS,
Najm MZ, Deo SV, Shukla NK and Husain SA: A study on genetic
variants of fibroblast growth factor receptor 2 (FGFR2) and the
risk of breast cancer from North India. PLoS One. 9:e1104262014.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Shan J, Dsouza SP, Bakhru S, Al-Azwani EK,
Ascierto ML, Sastry KS, Bedri S, Kizhakayil D, Aigha II, Malek J,
et al: TNRC9 downregulates BRCA1 expression and promotes breast
cancer aggressiveness. Cancer Res. 73:2840–2849. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Mcinerney N, Colleran G, Rowan A, Walther
A, Barclay E, Spain S, Jones AM, Tuohy S, Curran C, Miller N, et
al: Low penetrance breast cancer predisposition SNPs are site
specific. Breast Cancer Res Treat. 117:151–159. 2009. View Article : Google Scholar
|
|
21
|
Chalabi N, Bernard-Gallon DJ, Bignon YJ;
Breast Med Consortium; Kwiatkowski F, Agier M, Vidal V,
Laplace-Chabaud V, Sylvain-Vidal V, Bertholet V, et al: Comparative
clinical and transcriptomal profiles of breast cancer between
French and South Mediterranean patients show minor but
significative biological differences. Cancer Genomics Proteomics.
5:253–261. 2008.
|
|
22
|
Huijts PE, Vreeswijk MP, Kroeze-Jansema
KH, Jacobi CE, Seynaeve C, Krol-Warmerdam EM, Wijers-Koster PM,
Blom JC, Pooley KA, Klijn JG, et al: Clinical correlates of
low-risk variants in FGFR2, TNRC9, MAP3K1, LSP1 and 8q24 in a Dutch
cohort of incident breast cancer cases. Breast Cancer Res.
9:R782007. View
Article : Google Scholar : PubMed/NCBI
|
|
23
|
Sueta A, Ito H, Kawase T, Hirose K, Hosono
S, Yatabe Y, Tajima K, Tanaka H, Iwata H, Iwase H and Matsuo K: A
genetic risk predictor for breast cancer using a combination of
low-penetrance polymorphisms in a Japanese population. Breast
Cancer Res Treat. 132:711–721. 2012. View Article : Google Scholar
|
|
24
|
Ruiz-Narváez EA, Rosenberg L, Cozier YC,
Cupples LA, Adams-Campbell LL and Palmer JR: Polymorphisms in the
TOX3/LOC643714 locus and risk of breast cancer in African-American
women. Cancer Epidemiol Biomarkers Prev. 19:1320–1327. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Long J, Shu XO, Cai Q, Gao YT, Zheng Y, Li
G, Li C, Gu K, Wen W, Xiang YB, et al: Evaluation of breast cancer
susceptibility loci in Chinese women. Cancer Epidemiol Biomarkers
Prev. 19:2357–2365. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Pham TT, Angus SP and Johnson GL: MAP3K1:
Genomic alterations in cancer and function in promoting cell
survival or apoptosis. Genes Cancer. 4:419–426. 2013. View Article : Google Scholar
|
|
27
|
Hirschhorn JN, Lohmueller K, Byrne E and
Hirschhorn K: A comprehensive review of genetic association
studies. Genet Med. 4:45–61. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kim HC, Lee JY, Sung H, Choi JY, Park SK,
Lee KM, Kim YJ, Go MJ, Li L, Cho YS, et al: A genome-wide
association study identifies a breast cancer risk variant in ERBB4
at 2q34: Results from the Seoul breast cancer study. Breast Cancer
Res. 14:R562012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Barnholtz-Sloan JS, Shetty PB, Guan X,
Nyante SJ, Luo J, Brennan DJ and Millikan RC: FGFR2 and other loci
identified in genome-wide association studies are associated with
breast cancer in African-American and younger women.
Carcinogenesis. 31:1417–1423. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zheng Y, Ogundiran TO, Falusi AG,
Nathanson KL, John EM, Hennis AJ, Ambs S, Domchek SM, Rebbeck TR,
Simon MS, et al: Fine mapping of breast cancer genome-wide
association studies loci in women of African ancestry identifies
novel susceptibility markers. Carcinogenesis. 34:1520–1528. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Chen F, Chen GK, Millikan RC, John EM,
Ambrosone CB, Bernstein L, Zheng W, Hu JJ, Ziegler RG, Deming SL,
et al: Fine-mapping of breast cancer susceptibility loci
characterizes genetic risk in African Americans. Hum Mol Genet.
20:4491–4503. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Fu F, Wang C, Huang M, Song C, Lin S and
Huang H: Polymorphisms in second intron of the FGFR2 gene are
associated with the risk of early-onset breast cancer in Chinese
Han women. Tohoku J Exp Med. 226:221–229. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Maniatis T, Fritsch EF and Sambrook J:
1982 Molecular cloning: A laboratory manual. Cold Spring Harbor
Laboratory; Cold Spring Harbor, New York:
|
|
34
|
Hirotsu N, Murakami N, Kashiwagi T, Ujiie
K and Ishimaru K: Protocol: A simple gel-free method for SNP
genotyping using allele-specific primers in rice and other plant
species. Plant Methods. 6:122010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Li Z, Zhang Z, He Z, Tang W, Li T, Zeng Z,
He L and Shi Y: A partition-ligation-combination-subdivision EM
algorithm for haplotype inference with multiallelic markers: Update
of the SHEsis (http://analysis.bio-x.cn).
Cell Res. 19:519–523. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Haffty BG, Buchholz TA and Perez CA: Early
stage breast cancer. Principles and Practice of Radiation Oncology.
Halperin EC, Perez CA and Brady LW: 5th ed. J. B. Lippincott &
Co.; Philadelphia: pp. 1175–1291. 2008
|
|
37
|
Liang J, Chen P, Hu Z, Zhou X, Chen L, Li
M, Wang Y, Tang J, Wang H and Shen H: Genetic variants in
fibroblast growth factor receptor 2 (FGFR2) contribute to
susceptibility of breast cancer in Chinese women. Carcinogenesis.
29:2341–2346. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kawase T, Matsuo K, Suzuki T, Hiraki A,
Watanabe M, Iwata H, Tanaka H and Tajima K: FGFR2 intronic
polymorphisms interact with reproductive risk factors of breast
cancer: Results of a case control study in Japan. Int J Cancer.
125:1946–1952. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Stacey SN, Manolescu A, Sulem P,
Thorlacius S, Gudjonsson SA, Jonsson GF, Jakobsdottir M,
Bergthorsson JT, Gudmundsson J, Aben KK, et al: Common variants on
chromosome 5p12 confer susceptibility to estrogen receptor-positive
breast cancer. Nat Genet. 40:703–706. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Slattery ML, Baumgartner KB, Giuliano AR,
Byers T, Herrick JS and Wolff RK: Replication of five
GWAS-identified loci and breast cancer risk among Hispanic and
non-Hispanic white women living in the Southwestern United States.
Breast Cancer Res Treat. 129:531–539. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zheng W, Cai Q, Signorello LB, Long J,
Hargreaves MK, Deming SL, Li G, Li C, Cui Y and Blot WJ: Evaluation
of 11 breast cancer susceptibility loci in African-American women.
Cancer Epidemiol Biomarkers Prev. 18:2761–2764. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Shan J, Mahfoudh W, Dsouza SP, Hassen E,
Bouaouina N, Abdelhak S, Benhadjayed A, Memmi H, Mathew RA, Aigha
II, et al: Genome-wide association studies (GWAS) breast cancer
susceptibility loci in Arabs: Susceptibility and prognostic
implications in Tunisians. Breast Cancer Res Treat. 135:715–724.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Huijts PE, van Dongen M, de Goeij MC, van
Moolenbroek AJ, Blanken F, Vreeswijk MP, de Kruijf EM, Mesker WE,
van Zwet EW, Tollenaar RA, et al: Allele-specific regulation of
FGFR2 expression is cell type-dependent and may increase breast
cancer risk through a paracrine stimulus involving FGF10. Breast
Cancer Res. 13:R722011. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Raskin L, Pinchev M, Arad C, Lejbkowicz F,
Tamir A, Rennert HS, Rennert G and Gruber SB: FGFR2 is a breast
cancer susceptibility gene in Jewish and Arab Israeli population.
Cancer Epidemiol Biomarkers Prev. 17:1060–1065. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Broeks A, Schmidt MK, Sherman ME, Couch
FJ, Hopper JL, Dite GS, Apicella C, Smith LD, Hammet F, Southey MC,
et al: Low penetrance breast cancer susceptibility loci are
associated with specific breast tumor subtypes: Findings from the
breast cancer association consortium. Hum Mol Genet. 20:3289–3303.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Chen MB, Wu XY, Shen W, Wei MX, Li C, Cai
B, Tao GQ and Lu PH: Association between polymorphisms of
trinucleotide repeat containing 9 gene and breast cancer risk:
Evidence from 62,005 subjects. Breast Cancer Res Treat.
126:177–183. 2011. View Article : Google Scholar
|
|
47
|
He X, Yao G, Li F, Li M and Yang X:
Risk-association of five SNPs in TOX3/LOC643714 with breast cancer
in southern China. Int J Mol Sci. 15:2130–2141. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Garcia-Closas M, Hall P, Nevanlinna H,
Pooley K, Morrison J, Richesson DA, Bojesen SE, Nordestgaard BG,
Axelsson CK, Arias JI, et al: Heterogeneity of breast cancer
associations with five susceptibility loci by clinical and
pathological characteristics. PLoS Genet. 4:e10000542008.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Hunter DJ, Kraft P, Jacobs KB, Cox DG,
Yeager M, Hankinson SE, Wacholder S, Wang Z, Welch R, Hutchinson A,
et al: A genome-wide association study identifies alleles in FGFR2
associated with risk of sporadic postmenopausal breast cancer. Nat
Genet. 39:870–874. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
50
|
Shan J, Dsouza SP, Bakhru S, Al-Azwani EK,
Ascierto ML, Sastry KS, Bedri S, Kizhakayil D, Aigha II, Malek J,
et al: TNRC9 downregulates BRCA1 expression and promotes breast
cancer aggressiveness. Cancer Res. 73:2840–2849. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Chalabi N, Bernard-Gallon DJ, Bignon YJ,
Kwiatkowski F, Agier M, Vidal V, Laplace-Chabaud V, Sylvain-Vidal
V, Bertholet V, De Longueville F, et al: Comparative clinical and
transcriptomal profiles of breast cancer between French and South
Mediterranean patients show minor but significative biological
differences. Cancer Genomics Proteomics. 5:253–261. 2008.
|