Introduction
Thyroid cancer is the most common type of endocrine
cancer worldwide and accounts for the majority of endocrine
cancer-associated mortalities, although it represents only 1% of
all malignant diseases (1,2). Thyroid cancer-associated mortality
results from tumor invasion with local and distant metastases
(3). The following are at greater
risk for developing thyroid cancer: Caucasian individuals; females;
those with a low intake of iodine; and those that have previously
been exposed to radiation (4).
Furthermore, thyroid cancers are divided into three cancer
categories; differentiated, medullary and anaplastic (5). The treatments for all types of
anaplastic cancers include surgery, radioactive iodine treatment,
thyroid hormone supplementation, external beam radiation therapy
and chemotherapy (6).
Radiotherapy enables highly conformal treatment for
thyroid cancer and has been demonstrated to improve overall
survival (7,8). In a previous study, external
radiotherapy was administered to the thyroid bed and bilateral
cervical lymphatics as part of initial therapeutic treatment
(9). Furthermore, radiotherapy is
often performed in combination with enzyme or gene inhibitors, or
RNA interference (10).
Radiotherapy was reported to be enhanced by cyclooxygenase-2 enzyme
inhibitors (10). Radiotherapy was
performed with AZD2171 (a highly potent, orally active inhibitor of
vascular endothelial growth factor receptor signaling) to treat
lung cancer, and the result indicated that AZD2171 enhanced the
antitumor effects of radiotherapy (11). It has also been used in combination
with histone acetyltransferase and histone deacetylase (HDACs)
inhibitors for the treatment of prostate, esophageal, and head and
neck cancer (12).
Aberrant regulation of gene expression, attributed
to alterations in HDAC recruitment and activity, has consistently
been observed in solid and hematologic tumors (13). Therefore, HDACs can be considered
as potential therapeutic targets for the treatment of human
malignancies (14). HDACs are
important in the maintenance and function of chromatin by
regulating the acetylation state of histones (15). HDAC inhibitors are currently being
evaluated as anticancer agents in clinical trials, which can induce
growth arrest, differentiation and apoptosis of cancer cells in
in vitro and in vivo tumor-bearing animal models
(16–19). Trichostatin A (TSA) is a HDAC
inhibitor and inhibits growth of small cell lung cancer cells
(20). TSA may be administered
therapeutically for the redifferentiation of thyroid cancers to
promote radioiodide uptake (21).
Double-stranded RNA-mediated interference (RNAi) has recently
emerged as a notable tool in reverse genetic to silence gene
expression in multiple types of organism, including plants,
Caenorhabditis elegans and Drosophila (22). Furthermore, RNAi via the expression
of shRNA molecules is considered to be a particularly promising
tool in reverse genetics in mice, as it may enable inexpensive and
rapid gene function analysis in vivo (23).
In the present study, reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
analysis was performed to screen the enzymes that are associated
with the radiosensitivity of SW579 thyroid cancer cells. In
addition, to evaluate the potential impact of the screened enzymes
on SW579 cell radiosensitivity, shRNA-HDAC1, shRNA-HDAC2,
shRNA-HDAC4, shRNA-HDAC6 plasmids were constructed and shRNA pools
of the four shRNA plasmids were established. The cancer cells were
transfected with shRNA pools and irradiated using X-rays. The
morphology of cancer cells following radiotherapy was observed by
fluorescence microscopy.
Materials and methods
Plasmid construction
Using HDAC1, HDAC2, HDAC4 and HDAC6 mRNA sequences
that were obtained from GenBank (www.ncbi.nlm.nih.gov/genbank/) and shRNA design
principles, as described in a previous study (22), the online design software (siRNA
Selection Program; http://sirna.wi.mit.edu/), Ambion company, was used to
establish the target sequences. Four shRNA plasmid sequences
(shRNA-HDAC1, shRNA-HDAC2, shRNA-HDAC4 and shRNA-HDAC6) targeting
different coding regions of human HDAC1, HDAC2, HDAC4 and HDAC6
mRNA were designed. In addition, four scramble sequence shRNA
duplexes were synthesized to serve as negative controls (NCs) and
were abbreviated as shRNA-HDAC1-NC, shRNA-HDAC2-NC, shRNA-HDAC4-NC
and shRNA-HDAC6-NC. The sequences are presented in Table I. The shRNA pool, which contained
shRNA-HDAC1, shRNA-HDAC2, shRNA-HDAC4 and shRNA-HDAC6 were
constructed. The shRNA NC pool, which contained shRNA-HDAC1-NC,
shRNA-HDAC2-NC, shRNA-HDAC4-NC and shRNA-HDAC6-NC was also
established.
 | Table IThe mRNA sequence of different
shRNA-HDAC short fragment. |
Table I
The mRNA sequence of different
shRNA-HDAC short fragment.
| Index | Primer
sequence |
|---|
| shRNA-HDAC1-1 | F:
5′-CACCGCTCCATCCGTCCAGATAACACGAATGTTATCTGGACGGATGGAGC-3′ |
| R:
5′-AAAAGCTCCATCCGTCCAGATAACATTCGTGTTATCTGGACGGATGGAGC-3′ |
| shRNA-HDAC1-2 | F:
5′-CACCGCAAGCAGATGCAGAGATTCACGAATGAATCTCTGCATCTGCTTGC-3′ |
| R:
5′-AAAAGCAAGCAGATGCAGAGATTCATTCGTGAATCTCTGCATCTGCTTGC-3′ |
| shRNA-HDAC1-NC | F:
5′-CACCGCAATTTTTTTTTTTGATTCACGAATGAATCAAAAAAAAAAAAATGC-3′ |
| R:
5′-AAAAGCAATTTTTTTTTTTGATTCATTCGTGAATCAAAAAAAAAAAAATGC-3′ |
| shRNA-HDAC2-1 | F:
5′-CACCGCAGATGCAGAGATTTAATGTCGAAACATTAAATCTCTGCATCTGC-3′ |
| R:
5′-AAAAGCAGATGCAGAGATTTAATGTTTCGACATTAAATCTCTGCATCTGC-3′ |
| shRNA-HDAC2-2 | F:
5′-CACCGGCTGGAGGATTACATCATGCCGAAGCATGATGTAATCCTCCAGCC-3′ |
| R:
5′-AAAAGGCTGGAGGATTACATCATGCTTCGGCATGATGTAATCCTCCAGCC-3′ |
| shRNA-HDAC2-NC | F:
5′-CACCGCAATTTTTTTTTTTGATTCACGAATGAATCAAAAAAAAAAAAATGC-3′ |
| R:
5′-AAAAGCAATTTTTTTTTTTGATTCATTCGTGAATCAAAAAAAAAAAAATGC-3′ |
| shRNA-HDAC4-1 | F:
5′-CACCGGAGCTCGTGGTACTCAAACACGAATGTTTGAGTACCACGAGCTCC-3′ |
| R:
5′-AAAAGGAGCTCGTGGTACTCAAACATTCGTGTTTGAGTACCACGAGCTCC-3′ |
| shRNA-HDAC4-2 | F:
5′-CACCGGAACACATCAAGCACCAACACGAATGTTGGTGCTTGATGTGTTCC-3′ |
| R:
5′-AAAAGGAACACATCAAGCACCAACATTCGTGTTGGTGCTTGATGTGTTCC-3′ |
| shRNA-HDAC4-NC | F:
5′-CACCGCAATTTTTTTTTTTGATTCACGAATGAATCAAAAAAAAAAAAATGC-3′ |
| R:
5′-AAAAGCAATTTTTTTTTTTGATTCATTCGTGAATCAAAAAAAAAAAAATGC-3′ |
| shRNA-HDAC6-1 | F:
5′-CACCGCAATGGAAGAAGACCTAATCCGAAGATTAGGTCTTCTTCCATTGC-3′ |
| R:
5′-AAAAGCAATGGAAGAAGACCTAATCTTCGGATTAGGTCTTCTTCCATTGC-3′ |
| shRNA-HDAC6-2 | F:
5′-CACCGGAAGAGCTGATGTTGGTTCACGAATGAACCAACATCAGCTCTTCC-3′ |
| R:
5′-AAAAGGAAGAGCTGATGTTGGTTCATTCGTGAACCAACATCAGCTCTTCC-3′ |
| siRNA-HDAC6-NC | F:
5′-CACCGCAATTTTTTTTTTTGATTCACGAATGAATCAAAAAAAAAAAAATGC-3′ |
| R:
5′-AAAAGCAATTTTTTTTTTTGATTCATTCGTGAATCAAAAAAAAAAAAATGC-3′ |
| β-actin | F:
5′-GTGGACATCCGCAAAGAC-3′ |
| R:
5′-AAAGGGTGTAACGCAACTA-3′ |
Cell culture and plasmid
transfection
The SW579 human thyroid cancer cells were obtained
from the Shanghai Institutes for Biological Sciences, Chinese
Academy of Sciences (Shanghai, China). They were cultured in 10 ml
Dulbecco's modified Eagle's medium (DMEM; Thermo Fisher Scientific,
Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum
(FBS; Thermo Fisher Scientific, Inc.). Prior to transfection (12
h), ~2×105 cells/ml cells were seeded in 6-well plates
and 2 ml culture medium (90% DMEM and 10% FBS) was added to each
well. Cells were grown overnight to 50–60% confluence. All plasmids
were transfected with 8 µl metafectamine transfection
reagent (Biontex Laboratories GmbH, Martinsried/Planegg, Germany)
according to the manufacturer's instructions. shRNA expression
plasmid (1 µg) and 8 µl metafectamine were diluted in
serum- and antibiotic-free OptiMEM medium (50 µl; Thermo
Fisher Scientific, Inc.). The plasmid and metafectamine solutions
were combined and mixed by careful pipetting. Then the mixture was
incubated at room temperature for 20 min and added to the wells
containing the cells. The optiMEM was replaced with 2 ml DMEM and
10% FBS following incubation for 6 h. Cells were cultured in a
humidified atmosphere containing 5% CO2 at 37°C for 72
h. All transfection experiments were performed a minimum of three
times. To analyze the downregulation of HDACs by shRNA, three
groups were formed as follows: shRNA-HDAC1-1 (shRNA-HDAC2-1,
shRNA-HDAC4-1 and shRNA-HDAC6-1); shRNA-HDAC1-2 (shRNA-HDAC2-2,
shRNA-HDAC4-2 and shRNA-HDAC6-2); and shRNA-HDAC1-NC
(shRNA-HDAC2-NC, shRNA-HDAC4-NC and shRNA-HDAC6-NC). To investigate
the enhancement effect of shRNA pools on radiosensitivity of
thyroid cancer cells, the samples were divided into four groups:
Control (untransfected); TSA (Sigma-Aldrich, St. Louis, MO, USA);
shRNA pool; and shRNA NC pool. The TSA group was differentiated
from the control group by the addition of 200 µg/l HDAC
inhibitor, TSA to the cell culture medium.
Irradiation of thyroid cancer cells
Log-phase tumor cells were seeded as single-cell
suspensions into 6-well plates and were left to adhere overnight.
The cells were irradiated with a single dose (8 Gy) of X-rays
(FCS320; Gulmay Medical Ltd., Camberley, UK), and a standard
colony-forming assay was performed, according to a previous study
(24), to determine the respective
surviving fractions. To estimate the sensitivity to radiotherapy,
the cells were exposed to 3 µM 5-fluorouracil
(Sigma-Aldrich) for 16 h before irradiation, as described by
Spitzner et al (25). The
relative mRNA levels were determined by comparing the values to
mRNA levels prior to radiotherapy.
RT-qPCR
The expression changes of SW579 human thyroid cancer
cell epigenetic enzymes before and after radiotherapy were analyzed
by RT-PCR. The mRNA expression level of HDAC1, HDAC2, HDAC4 and
HDAC6 in cancer cells after transfection with shRNA-HDAC1-1,
shRNA-HDAC1-2, shRNA-HDAC1-NC, shRNA-HDAC2-1, shRNA-HDAC2-2,
shRNA-HDAC2-NC, shRNA-HDAC4-1, shRNA-HDAC4-2, shRNA-HDAC4-NC,
shRNA-HDAC6-1, shRNA-HDAC6-2 and shRNA-HDAC6-NC was also analyzed
by RT-PCR.
Total RNA was isolated from the cultured cells using
a TRIzol kit (Invitrogen; Thermo Fisher Scientific, Inc.). The RNA
quality was assessed using the NanoDrop 1000 (Thermo Fisher
Scientific, Inc.). RT-PCR was performed using QuantiTect Primer
Assay (Qiagen GmbH, Hilden, Germany) and QuantiTect SYBR Green
RT-PCR Kit (Qiagen GmbH) on a LightCycler 480 Instrument (Roche
Diagnostics, Mannheim, Germany). The detection and quantification
was performed as follows: Reverse transcription, 55°C for 30 min;
initial activation,15 min at 95°C; 40 cycles of denaturation, 94°C
for 15 sec; annealing, 30 sec at 55°C; and extension, 30 sec at
72°C. Fluorescence data was collected at the extension step. The
relative expression of the target gene was determined using the
2−ΔCt method (26).
Western blot analysis
Western blot analysis was performed on five groups
of thyroid cancer cells as follows: i) Thyroid cancer cells before
radiotherapy; ii) thyroid cancer cells after radiotherapy; iii)
thyroid cancer cells transfected with shRNA-HDAC-1; iv) thyroid
cancer cells transfected with shRNA-HDAC-2; and v) thyroid cancer
cells transfected with shRNA-HDAC-NC. The cells were collected
after transfection for 72 h, and washed with cold
phosphate-buffered saline (PBS; Sangon Biotech Co., Ltd., Shanghai,
China) twice. The cells were lysed with 1 ml lysis buffer (10% SDS,
0.5% Bromophenol Blue, 50% glycerol, 5% β-mercaptoethanol and 250
mmol/l Tris-HCl; pH 6.8; all obtained from Sangon Biotech Co.,
Ltd.) and phenylmethylsulfonyl fluoride (Sangon Biotech Co., Ltd.).
The mixture was centrifuged at 198 × g for 5 min after boiling for
10 min and the supernatants were collected. The protein
concentrations were determined according to the Pierce™ BCA Protein
assay kit (Thermo Fisher Scientific, Inc.). The cell lysate was
separated by 12% SDS-PAGE (Sangon Biotech Co., Ltd.) at 80 V for 30
min, then 120 V for 100 min and transferred to nitrocellulose
membranes (Mai Bio Co., Ltd., Shanghai, China) at 150 mA for 1 h.
After blocking non-specific binding sites with 5% non-fat milk, the
membrane was incubated with primary antibodies for 1.5 h at room
temperature. The primary antibodies used in the experiment were
mouse monoclonal anti-APE1 (1:3,000; Abcam, Cambridge, MA, USA;
cat. no. ab194), rabbit polyclonal anti-HDAC1 (1:10,000; Abcam;
cat. no. ab7028), rabbit monoclonal anti-HDAC2 (1:2,000; Abcam;
cat. no. ab32117), rabbit polyclonal anti-HDAC4 (1:500; Abcam; cat.
no. ab1437), rabbit polyclonal anti-HDAC6 (1:200; Abcam; cat. no.
ab1440) and monoclonal anti-β-actin (1:5,000; Santa Cruz
Biotechnology, Inc., Dallas, TX, USA; cat. no. sc-69879)
antibodies. The membrane was washed with Tris-buffered saline and
Tween-20 (Sangon Biotech Co., Ltd.) three times, for 10 min each
time. The membrane was then incubated with horseradish peroxidase
(HRP)-conjugated goat anti-rabbit IgG (1:5,000; Santa Cruz
Biotechnology, Inc.; cat. no. sc-2054) and rabbit anti-mouse IgG
(1:5,000; Santa Cruz Biotechnology, Inc.; cat. no. sc-358920)
antibodies. Images of the membranes were captured (LAS-3000;
Fujifilm, Tokyo, Japan), macroscopic observation was used to
compare the western blotting results with an LAS-3000 and Multi
Gauge (version 3.0; Fujifilm).
Morphology
Morphological analysis was performed on the four
groups of thyroid cancer cells (thyroid cancer cells, cells
cultured in medium with TSA, cells transfected with the shRNA pool
and cells transfected with the shRNA NC pool). Cells were collected
by centrifugation following two washes with PBS. Carboxyfluorescein
(1:500; Beijing Fanbo Biochemicals Co., Ltd., Beijing, China) was
used to mark the cells. Prior to observation, the cells were washed
three times in PBS, agitated for 5 min, dried and mounted with 30%
glycerol. A fluorescence microscope (Q550CW; Leica Microsystems
GmbH, Wetzlar, Germany; magnification, ×10) with Cooke software
version 3.04 (cooke-viewer.software.informer.com) was used to
perform apoptosis analysis from total cell and living cell numbers
counted.
Statistical analysis
All data were expressed as the mean ± standard
deviation from a minimum of three separate experiments. The
differences between groups were analyzed using Student's t-test in
the SPSS 10.0 software (SPSS, Inc., Chicago, IL, USA). P<0.05
was considered to indicate a statistically significant
difference.
Results
Epigenetic enzyme expression level
changes after radiotherapy
RT-PCR was used to analyze the epigenetic enzyme
expression changes in SW579 human thyroid cancer cells following
radiotherapy. The results are presented in Fig. 1. For the majority of the enzymes,
the mRNA expression level after radiotherapy was higher compared
with before irradiation. The increase was particularly notable in
HDAC1, HDAC2, HDAC4 and HDAC6.
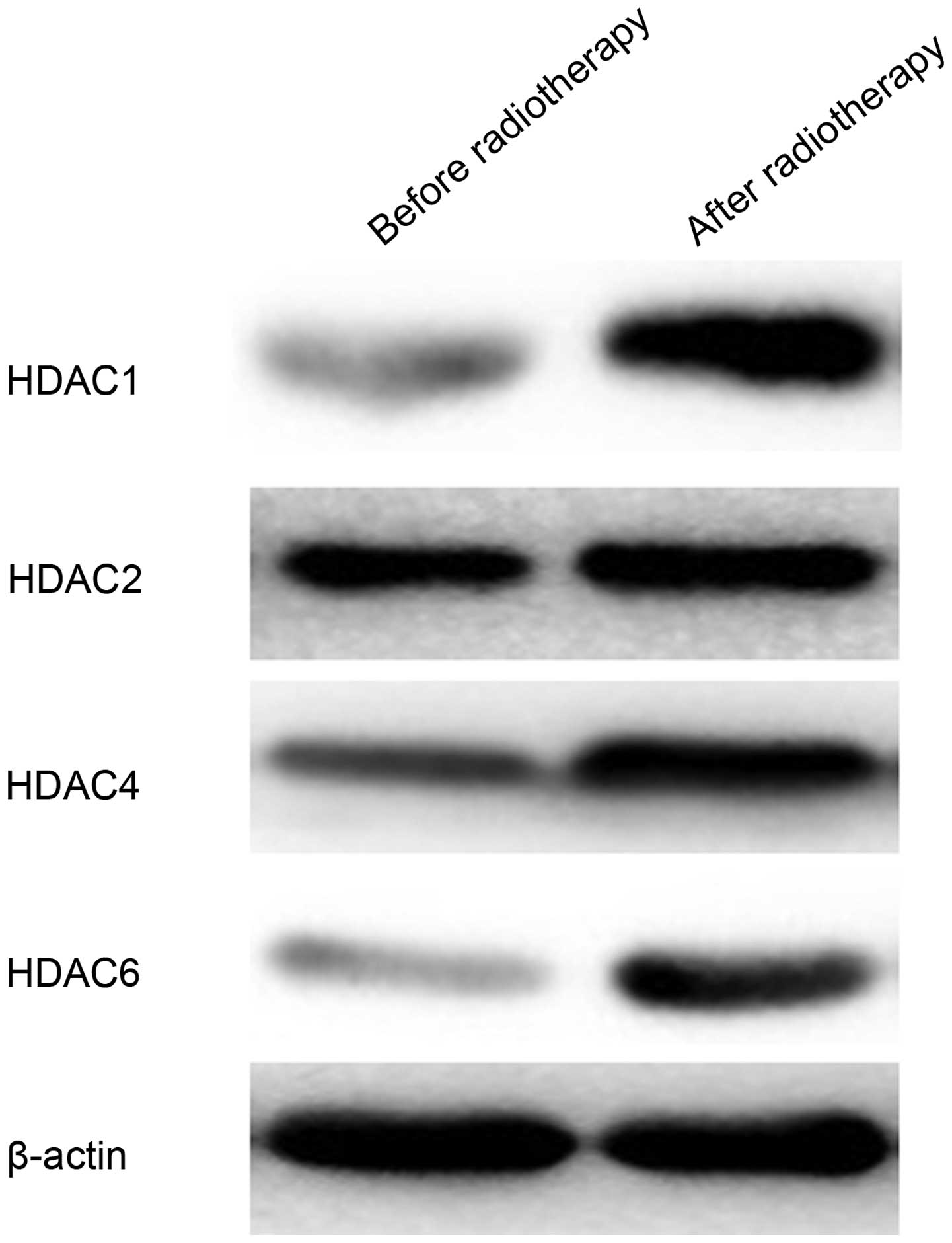
To further confirm the change in HDAC1, HDAC2, HDAC4
and HDAC6 protein expression, western blot analysis was performed
before and after radiotherapy (Fig.
2). The protein expression level of HDAC1, HDAC2 and HDAC6 in
cells after irradiation was markedly higher compared with before
irradiation. The protein expression intensity of HDAC4 was also
greater in cells after irradiation. These findings demonstrated
that radiotherapy upregulated the expression level of HDAC1, HDAC2,
HDAC4 and HDAC6 proteins.
Inhibition of HDAC expression after
transfection with shRNA-HDAC
ShRNA-HDAC1-1, shRNA-HDAC1-2, shRNA-HDAC1-NC,
shRNA-HDAC2-1, shRNA-HDAC2-2, shRNA-HDAC2-NC, shRNA-HDAC4-1,
shRNA-HDAC4-2, shRNA-HDAC4-NC, shRNA-HDAC6-1, shRNA-HDAC6-2 and
shRNA-HDAC6-NC plasmids were constructed, and transfected into
cultured SW579 thyroid cancer cells. To analyze the inhibition
effect of shRNA-HDAC plasmid on HDAC expression in the SW579 cells,
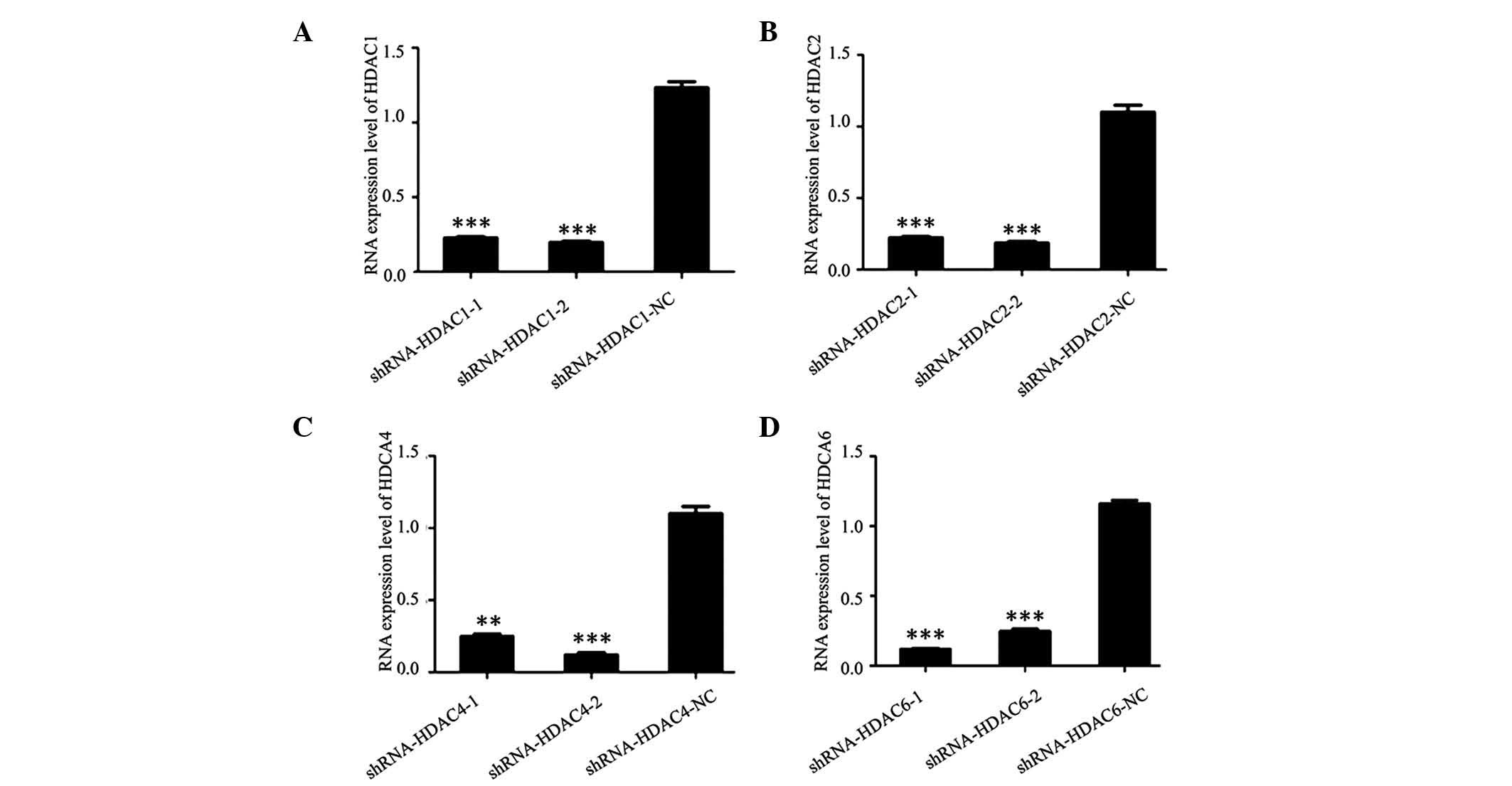
RT-PCR and western blot analysis were conducted (Fig. 3). As shown, the mRNA expression
level of HDAC1, HDAC2, HDAC4 and HDAC6 in thyroid cancer cells was
markedly decreased after transfection with shRNA-HDAC1,
shRNA-HDAC2, shRNA-HDAC4 and shRNA-HDAC6 plasmids, when compared
with those that were transfected with shRNA-HDAC1-NC,
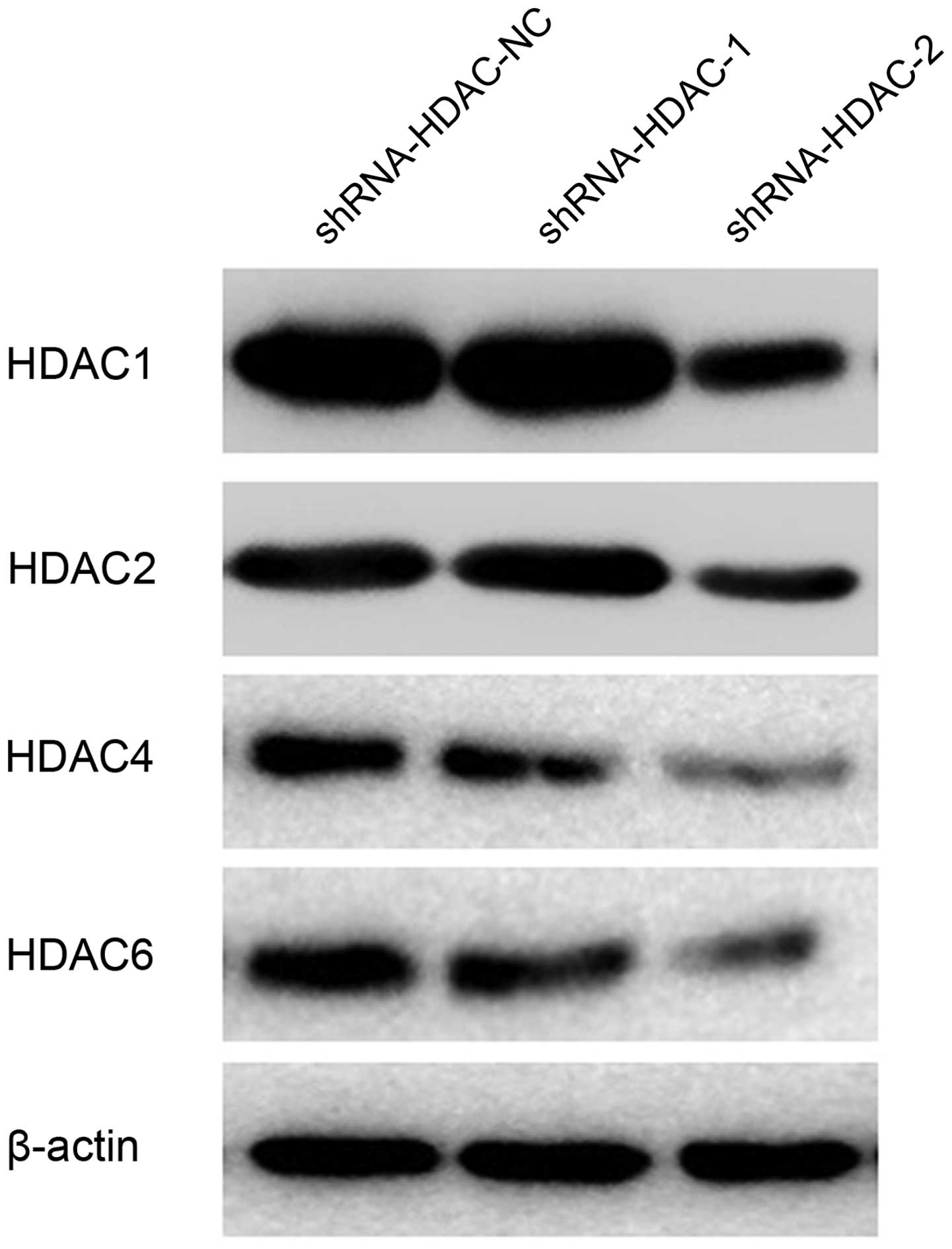
shRNA-HDAC2-NC, shRNA-HDAC4-NC and shRNA-HDAC6-NC (Fig. 4). The protein expression in the
positive interference group (shRNA-HDAC-1 and shRNA-HDAC-2) was
significantly weaker when compared with the negative interference
group (shRNA-HDAC-NC); the β-actin expression in the three groups
was similar.
Radiotherapy following transfection with
the shRNA pool
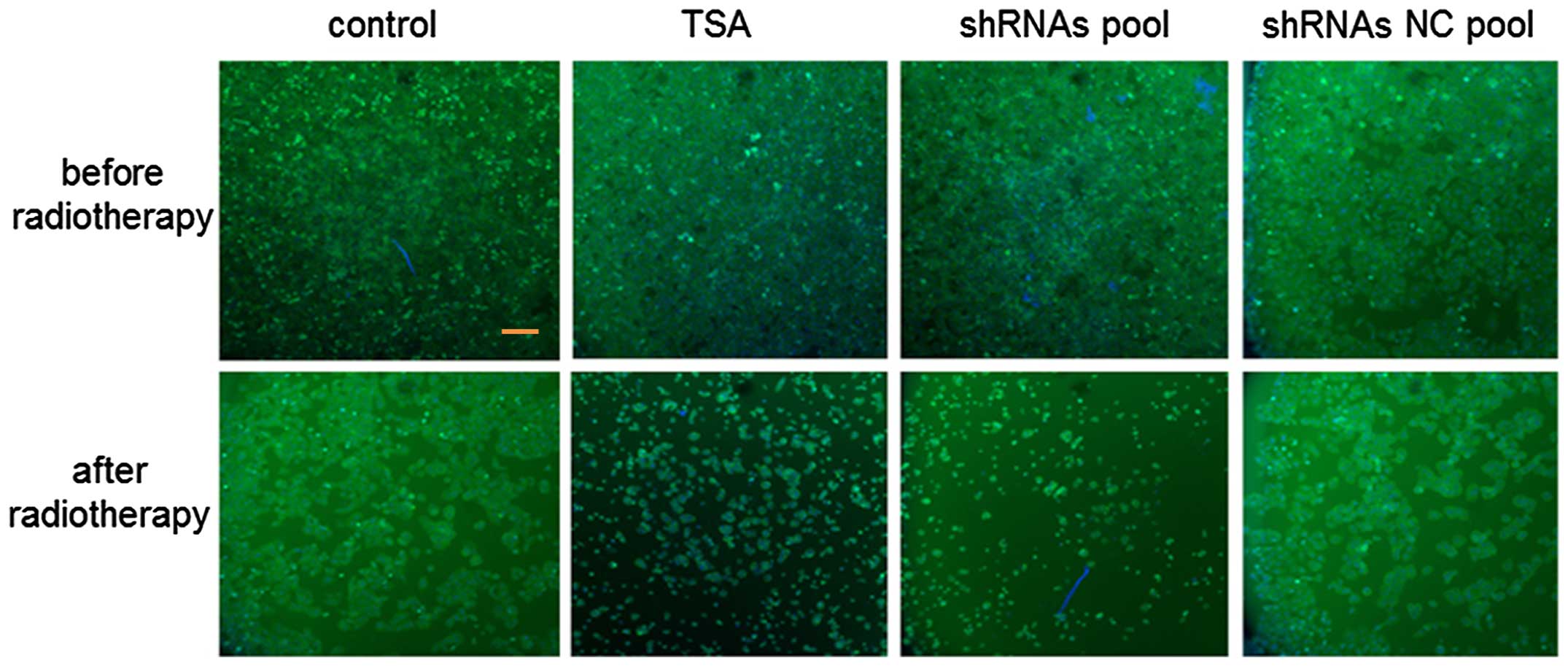
To investigate whether an shRNA pool plasmid
strengthens the sensitivity of SW579 cells to radiotherapy,
comparisons between the thyroid cancer cell morphology in the
control, TSA, shRNA pool and shRNAs NC pool groups was performed
(Fig. 5). The cell number in the
control and shRNA NC pool groups was notably higher than that of
the TSA and shRNA pool groups, which demonstrated that apoptosis
was promoted in the TSA and shRNA pool groups. Furthermore, the
cell number in the shRNA pool group was markedly lower than that of
the TSA group, indicating that the shRNA pool strengthened the
sensitivity of SW579 thyroid cancer cells to a greater extent than
the HDAC inhibitor, TSA.
Discussion
Thyroid cancer is associated with a high mortality
rate. Throughout the past two decades, an increasing incidence of
thyroid cancer has been reported worldwide (27). In the present study, RT-PCR was
used to analyze the epigenetic enzyme expression level changes in
SW579 thyroid cancer cells to investigate treatment of thyroid
cancer at the molecular level. The result demonstrated that the
expression level of HDAC1, HDAC2, HDAC4 and HDAC6 was markedly
increased following radiotherapy. It was hypothesized that HDAC1,
HDAC2, HDAC4 and HDAC6 expression may be associated with the
radiosensitivity of thyroid cancer cells and may be key in the
regulation of cellular radiosensitivity. Western blot analysis was
performed to further analyze the protein expression level of the
four enzymes, and the result indicated that the expression
intensity of HDAC1, HDAC2, HDAC4 and HDAC6 following radiotherapy
was markedly stronger than before radiotherapy. HDACs exert
antagonistic actions on histones, and regulate chromatin structure
and function by catalyzing removal of the acetyl modification from
the lysine residues of histones (28,29).
When HDACs act upon nucleosomal histones, it results in the tight
coiling of chromatin and silencing of the expression of various
genes, which include those implicated in the regulation of cell
survival, proliferation, differentiation, and apoptosis (30). HDAC1, HDAC2, HDAC3 and HDAC8 are
key in regulating cell proliferation and are expressed almost
exclusively in the nucleus of the majority of cell types (31). Overexpression of HDACs has been
reported in certain types of cancer tissue, such as stomach,
colorectal, breast, and lung (32–35).
Therefore, the present study proposed that HDAC1, HDAC2, HDAC4 and
HDAC6 were associated with the radiosensitivity of SW579 thyroid
cancer cells.
HDAC activity has been reported to be aberrant in
numerous types of cancer, therefore, HDAC inhibitors have been
adopted as potential anticancer therapies (36). Various HDAC inhibitors have been
developed and TSA is one example. In a previous study, TSA induced
apoptosis and cell-cycle arrest in anaplastic thyroid cancer cells
(37). The use of TSA to treat
thyroid cancer has been hypothesized to increase the efficacy of
radioactive iodine therapy (38).
Furthermore, TSA, when combined with radiotherapy, may induce cell
death in medulloblastoma (39). In
recent years, RNAi has developed rapidly and become an effective
and useful technique in molecular biology and oncology;
particularly regarding human gene function, signal transduction
research and gene therapy (40).
At present, RNAi is employed to downregulate the expression of
specific genes, which have been targeted in the gene therapy of
neurodegenerative diseases, Huntington's disease and the HIV
infection (41–43). In the present study, shRNA-HDAC1,
shRNA-HDAC1-NC, shRNA-HDAC2, shRNA-HDAC2-NC, shRNA-HDAC4,
shRNA-HDAC4-NC, shRNA-HDAC6 and shRNA-HDAC6-NC plasmids were
successfully constructed. Then, shRNA and shRNA NC pools of these
plasmids were established, and apoptosis analysis was performed on
four groups (control, TSA, shRNA pool and shRNA NC pool). In the
shRNA pool and shRNA NC pool groups, SW579 thyroid cancer cells
were transfected with the shRNA pool and shRNA NC pools,
respectively. The result demonstrated that the number of living
cells in the TSA and shRNA pool groups was markedly smaller than in
the control and shRNA NC pool groups. Furthermore, the number of
cells observed in the shRNA pool group was markedly lower when
compared with the TSA group. This finding demonstrated that TSA and
specific shRNA enhanced the sensitivity of SW579 thyroid cancer
cells to radiotherapy; furthermore, the effect of radiotherapy
enhanced by specific shRNA was greater than by TSA. In a previous
study, U251 glioma cells were transfected with shRNA-PGK1 and the
result indicated that tumor growth of the U251 xenografts was
significantly inhibited, which indicated the downregulation of PGK1
(resulting from transfection with shRNA) had sensitized the U251
xenografts to radiotherapy (44).
Wang et al (45) reported
that the silencing of signal transducer and activator of
transcription 3 expression using RNAi enhanced the efficacy of
radiotherapy for laryngeal carcinoma in vivo. In human
glioblastoma, the transfection of lentivirus-mediated shRNA to
glioblastoma cells was reported to inhibit NADPH oxidase 4, which
assisted with overcoming the radioresistance of cells and improved
its therapeutic efficacy for glioblastoma (46). In colorectal cancer, the silencing
of Wnt transcription factor, transcription factor 4 by specific
shRNA sensitized colorectal cancer cells to (chemo-) radiotherapy
(47). Therefore, the present
study concluded that silencing HDAC1, HDAC2, HDAC4 and HDAC6 using
an shRNA pool enhanced the sensitivity of SW579 thyroid cancer
cells to radiotherapy and the enhanced effect was more marked than
that of the HDAC inhibitor, TSA.
In conclusion, the enzymes, HDAC1, HDAC2, HDAC4 and
HDAC6 in SW579 thyroid cancer cells were sensitive to radiotherapy,
and their expression level was markedly increased following
radiotherapy. The transfection of an shRNA pool, which was
established using shRNA-HDAC1, shRNA-HDAC2, shRNA-HDAC4 and
shRNA-HDAC6 in the SW579 thyroid cancer cells, enhanced their
radiosensitivity. Thus, shRNA-targeted HDAC1, HDAC2, HDAC4 and
HDAC6 may present as a potential strategy for gene therapy of
thyroid cancer.
References
|
1
|
Mitsiades CS, Poulaki V, McMullan C, Negri
J, Fanourakis G, Goudopoulou A, Richon VM, Marks PA and Mitsiades
N: Novel histone deacetylase inhibitors in the treatment of thyroid
cancer. Clin Cancer Res. 11:3958–3965. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Abu-Amero KK, Alzahrani AS, Zou M and Shi
Y: High frequency of somatic mitochondrial DNA mutations in human
thyroid carcinomas and complex I respiratory defect in thyroid
cancer cell lines. Oncogene. 24:1455–1460. 2005. View Article : Google Scholar
|
|
3
|
Roomi MW, Bhanap B, Niedzwiecki A and Rath
M: Inhibitory effects of a novel nutrient mixture on MMP secretion
and invasion on human thyroid cancer cell line SW 579. JANA.
12:26–34. 2009.
|
|
4
|
McTiernan A, Weiss NS and Daling JR:
Incidence of thyroid cancer in women in relation to known or
suspected risk factors for breast cancer. Cancer Res. 47:292–295.
1987.PubMed/NCBI
|
|
5
|
Pasieka JL: Anaplastic thyroid cancer.
Curr Opin Oncol. 15:78–83. 2003. View Article : Google Scholar
|
|
6
|
O'Doherty MJ and Coakley AJ: Drug therapy
alternatives in the treatment of thyroid cancer. Drugs. 55:801–812.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Rosenbluth BD, Serrano V, Happersett L,
Shaha AR, Tuttle RM, Narayana A, Wolden SL, Rosenzweig KE, Chong LM
and Lee NY: Intensity-modulated radiation therapy for the treatment
of nonanaplastic thyroid cancer. Int J Radiat Oncol Biol Phys.
63:1419–1426. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wang Y, Tsang R, Asa S, Dickson B,
Arenovich T and Brierley J: Clinical outcome of anaplastic thyroid
carcinoma treated with radiotherapy of once-and twice-daily
fractionation regimens. Cancer. 107:1786–1792. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chow SM, Law SC, Mendenhall WM, Au SK,
Chan PT, Leung TW, Tong CC, Wong IS and Lau WH: Papillary thyroid
carcinoma: Prognostic factors and the role of radioiodine and
external radiotherapy. Int J Radiat Oncol Biol Phys. 52:784–795.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Choy H and Milas L: Enhancing radiotherapy
with cyclooxygenase-2 enzyme inhibitors: A rational advance? J Natl
Cancer Inst. 95:1440–1452. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Williams KJ, Telfer BA, Shannon AM, Babur
M, Stratford IJ and Wedge SR: Combining radiotherapy with AZD2171,
a potent inhibitor of vascular endothelial growth factor signaling:
pathophysiologic effects and therapeutic benefit. Mol Cancer Ther.
6:599–606. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Coucke P: A phase I trial on LBH 589
(panobinostat), a histone deacetylase inhibitor (HDAC) in
combination with external radiotherapy for the treatment of
prostate cancer, esophageal cancer and head and neck cancer. BJMO.
3:117–123. 2009.
|
|
13
|
Minucci S and Pelicci PG: Histone
deacetylase inhibitors and the promise of epigenetic (and more)
treatments for cancer. Nat Rev Cancer. 6:38–51. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Borbone E, Berlingieri MT, De Bellis F,
Nebbioso A, Chiappetta G, Mai A, Altucci L and Fusco A: Histone
deacetylase inhibitors induce thyroid cancer-specific apoptosis
through proteasome-dependent inhibition of TRAIL degradation.
Oncogene. 29:105–116. 2010. View Article : Google Scholar
|
|
15
|
Thurn KT, Thomas S, Moore A and Munster
PN: Rational therapeutic combinations with histone deacetylase
inhibitors for the treatment of cancer. Future Oncol. 7:263–283.
2011. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Gallinari P, Di Marco S, Jones P, Pallaoro
M and Steinkühler C: HDACs, histone deacetylation and gene
transcription: From molecular biology to cancer therapeutics. Cell
Res. 17:195–211. 2007.PubMed/NCBI
|
|
17
|
Giaginis C, Alexandrou P, Delladetsima I,
Giannopoulou I, Patsouris E and Theocharis S: Clinical significance
of histone deacetylase (HDAC)-1, HDAC-2, HDAC-4 and HDAC-6
expression in human malignant and benign thyroid lesions. Tumour
Biol. 35:61–71. 2014. View Article : Google Scholar
|
|
18
|
Shen L, Orillion A and Pili R: Histone
deacetylase inhibitors as immunomodulators in cancer therapeutics.
Epigenomics. 8:415–428. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang L, Bao X, Yang J, Li H, Zhou Q, Jiang
X, Li M, Liu X, Yuan X, Sun Y, et al: Novel cinnamohydroxamic acid
derivatives as HDAC inhibitors with anticancer activity in vitro
and in vivo. Chem Biol Interact. 249:64–70. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Platta CS, Greenblatt DY, Kunnimalaiyaan M
and Chen H: The HDAC inhibitor trichostatin A inhibits growth of
small cell lung cancer cells. J Surg Res. 142:219–226. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Clarke C, Burbridge E and Smyth P: Can
deacetylation promote radioiodide uptake in thyroid cancer?
Endocrine Abstracts. 9:1542005.
|
|
22
|
Sui G, Soohoo C, Affar el B, Gay F and Shi
Y, Forrester WC and Shi Y: A DNA vector-based RNAi technology to
suppress gene expression in mammalian cells. Proc Natl Acad Sci
USA. 99:5515–5520. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Seibler J, Küter-Luks B, Kern H, Streu S,
Plum L, Mauer J, Kühn R, Brüning JC and Schwenk F: Single copy
shRNA configuration for ubiquitous gene knockdown in mice. Nucleic
Acids Res. 33:e672005. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wu G, Ma Z, Qian J and Liu B: PP4R1
accelerates cell growth and proliferation in HepG2 hepatocellular
carcinoma. Onco Targets Ther. 8:2067–2074. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Spitzner M, Emons G, Kramer F, Gaedcke J,
Rave-Fränk M, Scharf JG, Burfeind P, Becker H, Beissbarth T,
Ghadimi BM, et al: A gene expression signature for
chemoradiosensitivity of colorectal cancer cells. Int J Radiat
Oncol Biol Phys. 78:1184–1192. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
27
|
Elashoff M, Matveyenko AV, Gier B,
Elashoff R and Butler PC: Pancreatitis, pancreatic and thyroid
cancer with glucagon-like peptide-1-based therapies.
Gastroenterology. 141:150–156. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kelly WK, O'Connor OA, Krug LM, Chiao JH,
Heaney M, Curley T, MacGregore-Cortelli B, Tong W, Secrist JP,
Schwartz L, et al: Phase I study of an oral histone deacetylase
inhibitor, suberoylanilide hydroxamic acid, in patients with
advanced cancer. J Clin Oncol. 23:3923–3931. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Luong QT, O'Kelly J, Braunstein GD,
Hershman JM and Koeffler HP: Antitumor activity of suberoylanilide
hydroxamic acid against thyroid cancer cell lines in vitro and in
vivo. Clin Cancer Res. 12:5570–5577. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Jones PA and Baylin SB: The fundamental
role of epigenetic events in cancer. Nat Rev Genet. 3:415–428.
2002.PubMed/NCBI
|
|
31
|
Khochbin S, Verdel A, Lemercier C and
Seigneurin-Berny D: Functional significance of histone deacetylase
diversity. Curr Opin Genet Dev. 11:162–166. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kim JH, Choi YK, Kwon HJ, Yang HK, Choi JH
and Kim DY: Downregulation of gelsolin and retinoic acid receptor
beta expression in gastric cancer tissues through histone
deacetylase 1. J Gastroenterol Hepatol. 19:218–224. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Giannini R and Cavallini A: Expression
analysis of a subset of coregulators and three nuclear receptors in
human colorectal carcinoma. Anticancer Res. 25:4287–4292.
2005.PubMed/NCBI
|
|
34
|
Krusche CA, Wülfing P, Kersting C, Vloet
A, Böcker W, Kiesel L, Beier HM and Alfer J: Histone deacetylase-1
and-3 protein expression in human breast cancer: A tissue
microarray analysis. Breast Cancer Res Treat. 90:15–23. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Sasaki H, Moriyama S, Nakashima Y,
Kobayashi Y, Kiriyama M, Fukai I, Yamakawa Y and Fujii Y: Histone
deacetylase 1 mRNA expression in lung cancer. Lung Cancer.
46:171–178. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Woyach JA, Kloos RT, Ringel MD, Arbogast
D, Collamore M, Zwiebel JA, Grever M, Villalona-Calero M and Shah
MH: Lack of therapeutic effect of the histone deacetylase inhibitor
vorinostat in patients with metastatic radioiodine-refractory
thyroid carcinoma. J Clin Endocrinol Metab. 94:164–170. 2009.
View Article : Google Scholar :
|
|
37
|
Furuya F, Shimura H, Suzuki H, Taki K,
Ohta K, Haraguchi K, Onaya T, Endo T and Kobayashi T: Histone
deacetylase inhibitors restore radioiodide uptake and retention in
poorly differentiated and anaplastic thyroid cancer cells by
expression of the sodium/iodide symporter thyroperoxidase and
thyroglobulin. Endocrinology. 145:2865–2875. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zarnegar R, Brunaud L, Kanauchi H, Wong M,
Fung M, Ginzinger D, Duh QY and Clark OH: Increasing the
effectiveness of radioactive iodine therapy in the treatment of
thyroid cancer using Trichostatin A, a histone deacetylase
inhibitor. Surgery. 132:984–990. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Kumar KS, Sonnemann J, Hong le TT, Buurman
C, Adler F, Maass M, Völker U and Beck JF: Histone deacetylase
inhibitors, but not vincristine, cooperate with radiotherapy to
induce cell death in medulloblastoma. Anticancer Res. 27:465–470.
2007.PubMed/NCBI
|
|
40
|
Wang J, Mi P, Lin G, Wang YX, Liu G and
Chen X: Imaging-guided delivery of RNAi for anticancer treatment.
Adv Drug Deliv Rev. Jan 22–2016.Epub ahead of print. View Article : Google Scholar
|
|
41
|
Bennasser Y, Yeung ML and Jeang KT: RNAi
therapy for HIV infection principles and practicalities. BioDrugs.
21:17–22. 2007. View Article : Google Scholar
|
|
42
|
Harper SQ: Progress and challenges in RNA
interference therapy for Huntington disease. Arch Neurol.
66:933–938. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Boudreau RL and Davidson BL: RNAi therapy
for neurodegenerative diseases. Curr Top Dev Biol. 75:73–92. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Cheng YJ, Ding H, Du HQ, Yan H, Zhao JB,
Zhang WB, Zou YJ, Liu HY and Xiao H: Downregulation of
phosphoglycerate kinase 1 by shRNA sensitizes U251 xenografts to
radiotherapy. Oncol Rep. 32:1513–1520. 2014.PubMed/NCBI
|
|
45
|
Wang HR, Li XM and Lu XY: Silencing of
signal transducer and activator of transcription 3 gene expression
using RNAi enhances the efficacy of radiotherapy for laryngeal
carcinoma in vivo. Zhonghua Er Bi Yan Hou Tou Jing Wai Ke Za Zhi.
44:591–596. 2009.In Chinese. PubMed/NCBI
|
|
46
|
Li Y, Han N, Yin T, Huang L, Liu S, Liu D,
Xie C and Zhang M: Lentivirus-mediated Nox4 shRNA invasion and
angiogenesis and enhances radiosensitivity in human glioblastoma.
Oxid Med Cell Longev. 2014:5817322014. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Kendziorra E, Ahlborn K, Spitzner M,
Rave-Fränk M, Emons G, Gaedcke J, Kramer F, Wolff HA, Becker H,
Beissbarth T, et al: Silencing of the Wnt transcription factor TCF4
sensitizes colorectal cancer cells to (chemo-) radiotherapy.
Carcinogenesis. 32:1824–1831. 2011. View Article : Google Scholar : PubMed/NCBI
|