Introduction
Borna disease virus (BDV), a neurotropic,
non-cytolytic, non-segmented RNA virus, is an enveloped virus of
~8.9 kb with six open reading frames (1,2),
which infects a wide variety of mammalian species, including
horses, sheep and dogs (3). BDV
has been widely investigated in neuroscientific fields on account
of its numerous unique attributes causing neurobehavioral diseases
(4) and the ability to introduce
its RNA transcripts into host genomes (5). Previous epidemiological studies have
shown that there may be a latent association between BDV infection
and human neuropsychiatric diseases (6), encephalitis and other brain diseases
(3,7–12).
In our previous study, BDV infection was reported in Chinese
neuropsychiatric patients and health care professionals (13,14),
which supported the hypothesis that BDV can infect humans and may
be a pathogen in certain mental disorders, although the underlying
molecular mechanism remains to be fully elucidated. However,
certain studies have found no direct evidence of BDV infection in
schizophrenia, bipolar disorder or major depressive disorder
(15–17). The controversy requires resolution
prior to use as a diagnostic method to ensure reliability.
BDV-associated functional disturbances of neuron and glial cells
have been evidenced (18–21) and its potential effects cannot be
ignored.
MicroRNAs (miRNAs) are small (~22 nucleotides in
length), non-coding, single-stranded RNAs (22). They regulate gene expression by
binding to the complementary sequence in the 3′-untranslated region
(3′-UTR) of target mRNAs, resulting in inhibited protein synthesis
or destabilizing of target mRNA translation at the
post-transcriptional level. miRNAs have been shown to be pervasive
in several biological processes, including cell death, cell
proliferation, the function of immune cells, hematopoiesis and
patterning of the nervous system (23). A wide range of studies have
revealed that miRNAs are associated with several human diseases,
including cancer, chronic inflammation and viral diseases (24–26).
Previous studies have also indicated that miRNAs have an effect on
neurodegenerative and neuropsychiatric disorders (27–30),
which suggests the possibility to associate the mechanisms of BDV
infection with miRNA dysregulation.
In the present study, miRNA arrays were used to
identify the differences in miRNA expression between
oligodendrocytes (OL cells) infected with the Hu-H1 BDV strain
(OL/BDV cells) and non-infected OL cells. The differentially
expressed miRNAs were then bioinformatically analyzed using Gene
Ontology (GO) and pathway analyses, in order to determine their
biological function and localization. Finally, reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
analysis was performed to validate the expression of the
differentially expressed miRNAs. The aim of the present study was
to determine which miRNAs are dysregualted in OL/BDV cells, and to
facilitate further investigation of the role of miRNAs in BDV
infection.
Materials and methods
Cell line and preparation of BDV Hu-H1
strain solution
The BDV Hu-H1 strain (passages 75–76 in OL cells),
originally isolated from PBMCs of a patient with bipolar disorder,
and a human fetal-derived OL cell line, were provided by Professor
Hanns Ludwig (Free University of Berlin, Berlin, Germany) (31). Dulbecco's modified Eagle's medium
(DMEM), fetal bovine serum (FBS), penicillin-streptomycin solution,
phosphate-buffered saline (PBS), 0.25% trypsin-EDTA and L-glutamine
were purchased from GE Healthcare Life Sciences (Logan, UT, USA).
The human OL cell line infected with the BDV Hu-H1 strain was
cultured with DMEM in 10% FBS and 100 U/ml penicillin/streptomycin
in a humidified incubator (5% CO2; 37°C). The preparation and viral
titration of the BDV Hu-H1 solution were performed, as described
previously (32). The cells in 20
10-cm dishes (density, 107) were washed twice with PBS,
and 1 ml fresh growth medium was added when the cells in the dishes
reached 90% confluence. The cell solution was then frozen (−80°C)
and thawed (25°C) for 15 min, and repeated three times. The lysate
was then centrifuged at 3,000 × g for 10 min at room temperature.
The resulting supernatant, which contained infectious viral
particles, was used as the stock viral solution.
The OL cells were seeded into 96-well plates
(3×104 cells/well). At 8 h post-adherence, the medium
was removed and 100 µl viral solutions were added to each well. The
stock viral solution was serially diluted 10-fold five times, with
four replicates for each concentration. The cells were cultured for
7 days in DMEM/2% FBS, during which the cell medium was replaced
once every 2 days to maintain the extracellular environment. The
viral titration was assessed using immunohistochemistry. The
BDV-infected OL cells were fixed in 96-well plates for 30 min at
room temperature with 4% paraformaldehyde, followed by
permeabilization for 10 min in 0.25% Triton X-100. The cells were
then rinsed three times with PBS (5 min each time) and blocked with
5% (w/v) skimmed milk solution for 1 h at 37°C. The cells were then
incubated overnight with mouse anti-BDV-specific nuclear-protein
(p40) antigen primary monoclonal antibody (provided by Professor
Ludwig Hanns, 1:1,000 diluted with PBS) (33) at 4°C, followed by incubation for 1
h with secondary goat anti-mouse antibody (cat. no. A0216; 1:5,000;
Beyotime Institute of Biotechnogy, Shanghai, China) at room
temperature. Immunofluorescence was detected using a phase-contrast
microscope following three PBS washes (32).
BDV infection of OL cells
A total of 105 non-infected OL cells were seeded
into four separate 6-well plates (total 24 wells) with 10% FBS in
DMEM. Half of these wells were infected with Hu-H1 stock solution
(as above) at a multiplicity of infection of 1.0. Specifically,
following adherence of the OL cells, the medium was removed, and
150 µl Hu-H1 strain solution was added per well to produce
BDV-infected OL (OL/BDV) cells. The cells were stored in a
humidified incubator (5% CO2 at 37°C) for 1.5–2 h. Next, the excess
viral solution was removed by suction at the edge of the plated,
and the cells were cultured in fresh medium. The remaining 12 wells
of OL cells were maintained as non-infected control OL cells. The
two cell groups were incubated under the same conditions for the
remainder of the experiment. The BDV infection was detected and
observed using an immunouorescence assay, as described previously
(32,34).
miRNA arrays
On day 14 post-infection, six wells of the OL and
OL/BDV cells were used for miRNA arrays, respectively. Fluorescent
miRNA targets were prepared from 1 or 2.5 µg total RNA samples,
which were extracted from the OL/BDV and non-infected OL cells
using an OneArray® Amino Allyl miRNA Amplification kit
(Phalanx Biotech Group, Hsinchu, Taiwan) and Cy5 dyes (GE
Healthcare, Piscataway, NJ, USA). Fluorescent targets were
hybridized to the Human Whole Genome OneArray® using a
Phalanx hybridization buffer on the Phalanx miRNA
OneArray® Hybridization system (Phalanx Biotech Group).
Following 16 h of hybridization at 50°C, non-specific binding
targets were removed through three washing steps (42°C for 5 min,
42°C for 5 min and 25°C for 5 min, followed by rinsing 20 times),
and the slides were dried by centrifugation at 1,000 × g for 3 min
at room temperature and scanned using an Axon 4000B scanner
(Molecular Devices LLC, Sunnyvale, CA, USA). The intensities of
each probe were obtained using GenePix 4.1 software (Molecular
Devices LLC). The probes with a log2 ratio ≥0.58 or ≤-0.58, and
P<0.05 were defined as differential genes for further pathway
enrichment analysis. Each experiment was repeated three times.
Prediction of target genes and
bioinformatic analysis
GO analysis (www.geneontology.org) was applied to determine the
functions of the intersecting genes on the basis of molecular
function, cellular component and biological process. To ensure
understanding of the gene expression information, the pathways
(www.reactome.org/ReactomeGWT/entrypoint.html and
www.genome.jp/kegg/) of the target genes
of the different miRNA were also analyzed.
RT-qPCR analysis
On day 14 post-infection, the remaining six wells of
OL and OL/BDV cells were used for RT-qPCR assays, respectively.
Total RNA was extracted using the miRNEasy Mini kit (cat.
no.217004; Qiagen, Hilden, Germany). RT-qPCR was performed on a
Corbett Research Rotor-Gene 6000 thermocycler (Corbett Life
sciences, Sydney, Australia). The All-in-OneTM First-Strand cDNA
Synthesis kit (cat. no. AOPT-0020) and All-in-One™ miRNA qPCR
Detection kit (cat. no. AOPR-0200) was purchased from GeneCopoeia,
Inc. (GeneCopoeia Inc., MD, USA). All 10 pairs of miRNA primers and
the U6 primers for the RT-qPCR were purchased from GeneCopoeia,
Inc. (GeneCopoeia, Inc. Guangzhou, China). Briefly, the volume used
for RT was 25 µl, comprising 5 µl 5X RT buffer, 1 µl 2.5 U/µl PolyA
polymerase, 1 µl RTase Mix, 2,000 ng total RNA template and
RNase-free water. RT was performed using a Gene Amp PCR system 9700
(Applied Biosystems Life Technologies; Thermo Fisher Scientific,
Inc., Waltham, MA, USA) at 37°C for 60 min and 85°C for 5 min.
The RT-qPCR assays used a total volume of 20 µl,
according to protocol, comprising 10 µl 2X All-in-One qPCR mix, 2
µl All-in-OneTM miRNAqPCR primer (2 µM), 2 µl Universal Adaptor PCR
primer (2 µM) and 2 µl cDNA (diluted 1:5). All reactions were run
in a Corbett Research Rotor-Gene 6000 thermocycler for 40 cycles,
which consisted of 94°C for 5 min, followed by 35 cycles of 94°C
for 30 sec, 58°C for 30 sec and 68°C for 30 sec. The melting
analysis of the PCR products was performed as follows: Temperature
was increased between 50 and 99°C (1°C increase at each step), with
a 90 sec period of pre-melt conditioning in the first step, and 5
sec for each subsequent step. Each experiment was repeated three
times. Values were normalized against the expression levels of U6,
and ΔΔCq values were calculated. The relative abundance of each
miRNA was calculated using the 2−ΔΔCq method (35–37).
Statistical analysis
For all miRNA quantification experiments,
quantification cycle (Cq) values >35 were excluded. Values were
normalized against the expression levels of U6, and ΔΔCq values
were calculated. Statistical analysis was performed using SPSS 19.9
software (IBM SPSS, Armonk, NY, USA). Student's t-test was
used to analyze the differences in miRNA expression between the
OL/BDV and non-infected OL cells. P<0.05 was considered to
indicate a statistically significant difference. All experiments
were repeated at least three times.
Results
miRNA expression profiling
To evaluate the different miRNAs between the OL/BDV
cells and non-infected OL cells, the present study profiled the
expression levels of 657 miRNAs in the two groups using an miRNA
array. Compared with the non-infected OL cells, a total of 10
miRNAs were differentially expressed in the OL/BDV cells (four
upregulated; six downregulated). miR-146a-5p showed the highest
expression level in the OL/BDV cells, whereas miR-4521 and
miR-3676-3p showed the lowest expression levels (Fig. 1).
GO and pathway analyses of miRNA
target genes
The target genes of the differentially expressed
miRNAs were predicted using the online database (http://targetscan.org/ and http://www.mirbase.org/), and then submitted for GO
functional classification and pathway analysis (http://www.broadinstitute.org/gsea/msigdb/annotate.jsp).
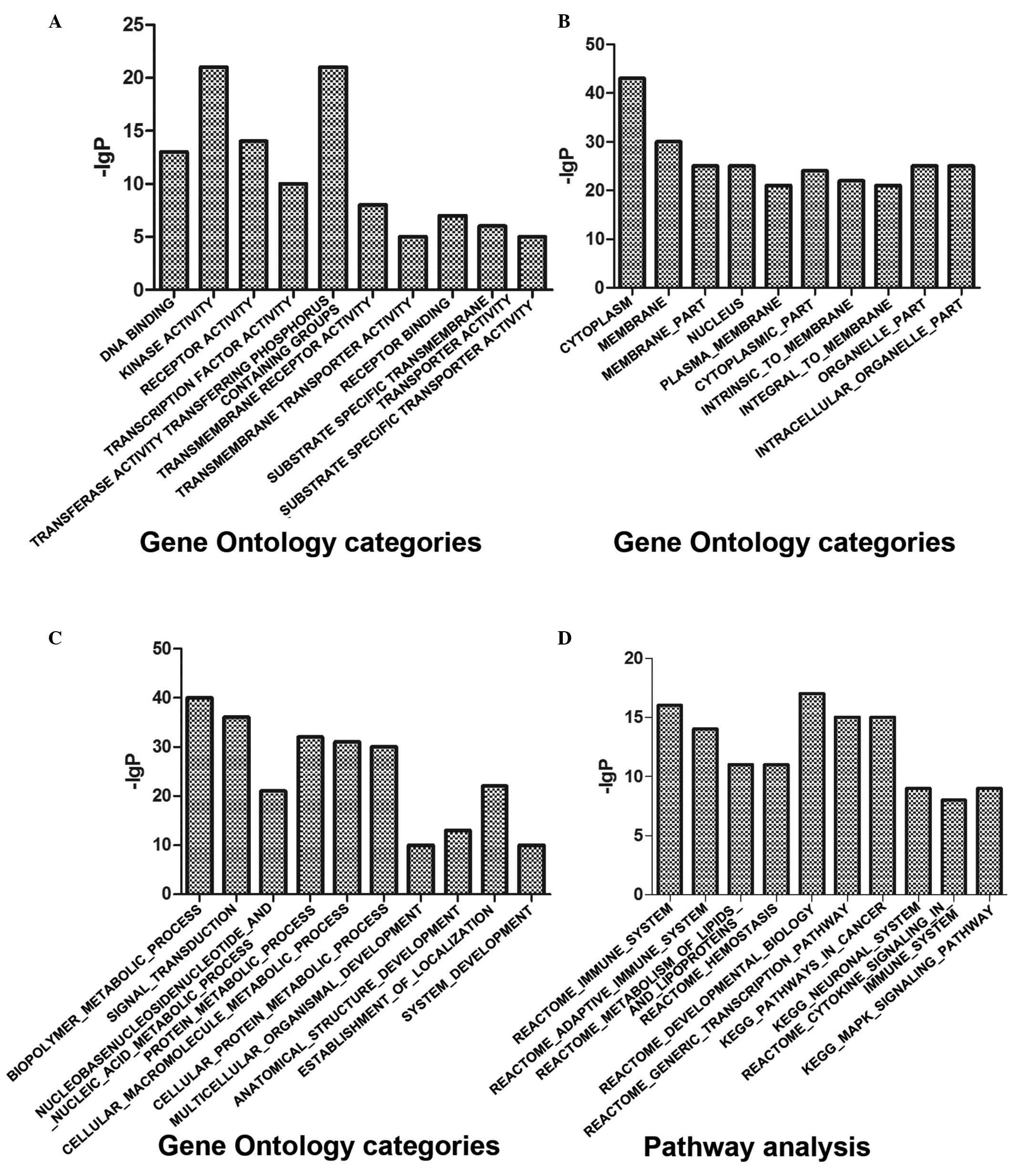
The molecular functions of the target genes were predominantly
associated with ‘DNA binding and receptor activity’, and involved
in certain biological process, including ‘biopolymer metabolic
process’ and ‘signal transduction’ (Fig. 2A-C; Table I). In terms of pathway analysis,
the immune system and adaptive immune system were the most
significant pathways (Fig. 2D;
Table I).
 | Table I.Gene Ontology analyses and pathway
analyses of microRNA target genes. |
Table I.
Gene Ontology analyses and pathway
analyses of microRNA target genes.
| Gene function | Genes (n) | P-value |
|---|
| Molecular
function |
|
|
| DNA
binding | 602 | 3.53E-13 |
|
Receptor activity | 583 | 8.94E-14 |
|
Transferase activity,
transferring phosphorus-containing groups | 424 | 6.75E-21 |
|
Transmembrane receptor
activity | 419 | 1.34E-08 |
|
Substrate-specific transporter
activity | 392 | 1.22E-05 |
| Cellular
component |
|
|
|
Cytoplasm | 2,131 | 1.88E-43 |
|
Membrane | 1,994 | 9.85E-30 |
|
Membrane part | 1,670 | 6.37E-25 |
|
Nucleus | 1,430 | 1.37E-25 |
| Plasma
membrane | 1,426 | 6.07E-21 |
| Biological
process |
|
|
|
Biopolymer metabolic
process | 1,684 | 1.98E-40 |
| Signal
transduction | 1,634 | 1.46E-36 |
|
Nucleobase, nucleoside,
nucleotide and nucleic acid metabolic process | 1,244 | 5.47E-21 |
| Protein
metabolic process | 1,231 | 1.89E-32 |
|
Cellular macromolecule
metabolic process | 1,131 | 6.78E-31 |
| Pathway
analyses |
|
|
| Immune
system | 933 | 5.06E-16 |
|
Adaptive immune system | 539 | 1.03E-14 |
|
Metabolism of lipids and
lipoproteins | 478 | 2.37E-11 |
|
Hemostasis | 466 | 3.29E-11 |
|
Developmental biology | 396 | 1.52E-17 |
Validation of differential miRNA
expression using RT-qPCR analysis
RT-qPCR analysis was performed to validate the
expression levels of the 10 differentially expressed miRNAs in the
OL/BDV cells. The relative expression levels of miRNAs were
normalized against the expression levels of U6. Of the 10 miRNAs,
seven exhibited significantly lower levels of expression in the
OL/BDV cells: miR-1908, miR-3676-3p, miR-296-3p, miR-146a-5p,
miR-1290, miR-424-5p and miR-7-5p (Fig. 3). No differences in expression were
found in miR-1244 or miR-4521 between the two groups (Fig. 3), and only one miRNA (miR-4433-3p)
was undetected in the RT-qPCR analysis. Of note, RT-qPCR showed
that the expression of miR-146a-5p, which was upregulated in the
miRNA array, was downregulated in the OL/BDV cells.
 | Figure 3.Validation of differential miRNA
expression using RT-qPCR analysis. Of the 10 miRNAs, seven
(miR-1908, miR-3676-3p, miR-296-3p, miR-146a-5p, miR-424-5p,
miR-7-5p and miR-1290) were expressed at significantly lower levels
in the BDV-infected OL (OL/BDV) cells, compared with the
non-infected OL cells. No significant differences were found in two
of the miRNAs (miR-1244 and miR-4521) between the two cell groups.
The remaining miRNA was not detected using RT-qPCR. Data are
expressed as the mean ± standard deviation. *P<0.01,
#P<0.05 and ∆P>0.05, vs. OL group.
miRNA/miR, microRNA; OL, oligodendrocyte; BDV, borna disease virus;
RT-qPCR, reverse transcription-quantitative polymerase chain
reaction. |
Discussion
BDV is a neurotropic virus, which can cause central
nervous system dysfunction in several mammalian species, including
humans (3,36). BDV Hu-H1, originally derived from a
human bipolar patient (31), can
induce apoptosis and metabolic dysfunction in human OL cells in
vitro (32,38). Proteomic analyses have indicated
that BDV Hu-H1 can activate the downstream extracellular
signal-regulated kinase (ERK)-ribosomal S6 kinase complex of the
Raf/mitogen-activated protein kinase (MAPK) kinase/ERK signaling
cascade in human OL cells (39).
Additionally, BDV Hu-H1 can result in brain metabolic dysfunction
in Sprague-Dawley rats (40).
Despite these findings, the mechanisms underlying BDV Hu-H1
infection in the human brain remain to be fully elucidated. miRNAs
may provide a novel approach to addressing remaining question.
Thus, the present study profiled and analyzed miRNA expression in
BDV Hu-H1-infected human OL cells.
An miRNA array is a high throughput and versatile
screening tool for analyzing the expression of miRNA, however, its
false positives cannot be ignored. Therefore, microarray data
requires validation using RT-qPCR analysis, which has higher
specificity and provides reliable quantity. It was reported in
previous studies that preliminary results were inconsistent with
the results of RT-qPCR analysis (41,42).
Similarly, in the present study, miR-146a-5p, miR-1290, miR-1908
and miR-424-5p showed downregulation in expression levels using
RT-qPCR analysis, which was inconsistent with the results of the
miRNA array. In addition, miR-4433-3p was not detected using
RT-qPCR analysis, and no differences were found in the expression
of miR-1244 or miR-4521 between the BDV-infected and non-infected
OL cells. However, three consistently downregulated miRNAs were
found: miR-7-5p, miR-296-3p and miR-3676-3p.
In the dysregulated miRNAs, the present study
focused on miR-7-5p, miR-424-5p and miR-296-3p, which are closely
associated with neural cell proliferation and apoptosis. miR-7-5p
has been reported in a wide range of signaling pathways, including
the MAPK and phosphoinositide 3-kinase (PI3K)/Akt pathway (43,44).
miR-7-5p can inhibit vascular endothelial cell proliferation via
directly targeting the 3′-UTR of RAF1, an upstream element of the
Ras-Raf-MAPK pathway, which has a key effect on nervous system
function. However, miR-7-5p is frequently downregulated in
glioblastoma microvasculature (43). miR-7 can efficiently affect cell
proliferation and metastasis in hepatocellular carcinoma, through
regulation of the PI3K/AKT pathway by suppressing PIK3CD, mammalian
target of rapamycin and p70S6 K (44). In addition, miRNA-7-5p can inhibit
melanoma cell migration and invasion by regulating insulin receptor
substrate-2 (45). The functions
of miR-7-5p have also been reported in other types of cancer by
targeting different signaling pathways (46–50).
These results provide novel information for further investigating
the function of miR-7-5p in BDV-infected nervous cells.
miR-424-5p is upregulated and modulates the ERK1/2
signaling pathway by targeting suppressor of cytokine induced
signaling 6 in pancreatic cancer (51). However, its downregulation can lead
to the progression of liver cancer, and regulate the resistance to
anoikis and epithelial mesenchymal transition during the metastatic
process of hepatocellular carcinoma cells by targeting inhibitor of
β-catenin and T cell factor (52).
In addition, previous studies have shown that activation of the
ERK1/2 pathway may hinder nerve growth factor-induced cell
differentiation in BDV-infected PC12 cells (19). Therefore, whether dysregulated
miR-424-5p affects BDV-infected nervous cells through the ERK1/2
signaling pathway requires further investigation. Although reports
of miR-296-3p are limited, miR-296-3p has been found to regulate
cell growth by targeting the potassium channel, EAG1 (53), in human glioblastoma, which
presents a novel view in understanding the pathogenic mechanisms of
BDV infection.
Previous studies have reported that human OL cells
infected with the BDV H1766 strain, isolated from a horse, revealed
downregulated expression levels of miR-122 and miR-155 (54,55),
which differed from the results of the BDV Hu-H1 infected and
non-infected OL cells in the present study. Of note, miRNA
expression profiling may be unique due to the possible divergent
mechanisms of human OL cells infected with different BDV strains;
this requires further investigation.
In conclusion, the present study screened 10 of 657
dysregulated miRNAs in BDV Hu-H1-infected human OL cells using an
miRNA array. Of the 10 miRNAs validated using RT-qPCR, the
expression levels of seven were significantly downregulated:
miR-1908, miR-3676-3p, miR-296-3p, miR-146a-5p, miR-424-5p,
miR-7-5p and miR-1290. The biological functions of the dysregulated
miRNAs require further investigation, however, based on GO and
pathway analyses, further investigation of BDV-infected OL cells
offers promise in improving current understanding of the
neuropathogenesis of BDV.
Acknowledgements
This study was supported by the National Natural
Science Foundation of China (grant no. 31300137), the National Key
Scientific Program of China (grant no. 2009CB918300) and the
Medical Scientific Research Project of Chongqing Health Bureau
(grant no 20142022). The authors would like to thank Professor Liv
Bode and Professor Hanns Ludwig of the Robert Koch Institute and
the Institute of Virology at the Free University of Berlin for
providing the human OL cell line and the BDV Hu-H1 strain used in
the present study. The authors would also like to thank the
scientific editors at Impactys (www.impactys.com) for their editing and proofreading
services.
References
|
1
|
Briese T, Schneemann A, Lewis AJ, Park YS,
Kim S, Ludwig H and Lipkin WI: Genomic organization of Borna
disease virus. Proc Natl Acad Sci USA. 91:4362–4366. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Cubitt B, Oldstone C and de la Torre JC:
Sequence and genome organization of Borna disease virus. J Virol.
68:1382–1396. 1994.PubMed/NCBI
|
|
3
|
Ludwig H and Bode L: Borna disease virus:
New aspects on infection, disease, diagnosis and epidemiology. Rev
Sci Tech. 19:259–288. 2000.PubMed/NCBI
|
|
4
|
Ludwig H, Bode L and Gosztonyi G: Borna
disease: A persistent virus infection of the central nervous
system. Prog Med Virol. 35:107–151. 1988.PubMed/NCBI
|
|
5
|
Horie M, Honda T, Suzuki Y, Kobayashi Y,
Daito T, Oshida T, Ikuta K, Jern P, Gojobori T, Coffin JM and
Tomonaga K: Endogenous non-retroviral RNA virus elements in
mammalian genomes. Nature. 463:84–87. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wang X, Zhang L, Lei Y, Liu X, Zhou X, Liu
Y, Wang M, Yang L, Zhang L, Fan S and Xie P: Meta-analysis of
infectious agents and depression. Sci Rep. 4:45302014.PubMed/NCBI
|
|
7
|
Bode L, Reckwald P, Severus WE, Stoyloff
R, Ferszt R, Dietrich DE and Ludwig H: Borna disease virus-specific
circulating immune complexes, antigenemia, and free antibodies-the
key marker triplet determining infection and prevailing in severe
mood disorders. Mol Psychiatry. 6:481–491. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bode L, Zimmermann W, Ferszt R, Steinbach
F and Ludwig H: Borna disease virus genome transcribed and
expressed in psychiatric patients. Nat Med. 1:232–236. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Fu ZF, Amsterdam JD, Kao M, Shankar V,
Koprowski H and Dietzschold B: Detection of Borna disease
virus-reactive antibodies from patients with affective disorders by
western immunoblot technique. J Affect Disord. 27:61–68. 1993.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Rott R, Herzog S, Fleischer B, Winokur A,
Amsterdam J, Dyson W and Koprowski H: Detection of serum antibodies
to Borna disease virus in patients with psychiatric disorders.
Science. 228:755–756. 1985. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sauder C, Müller A, Cubitt B, Mayer J,
Steinmetz J, Trabert W, Ziegler B, Wanke K, Mueller-Lantzsch N, de
la Torre JC and Grässer FA: Detection of Borna disease virus (BDV)
antibodies and BDV RNA in psychiatric patients: Evidence for high
sequence conservation of human blood-derived BDV RNA. J Virol.
70:7713–7724. 1996.PubMed/NCBI
|
|
12
|
Thakur R, Sarma S and Sharma B: Role of
Borna disease virus in neuropsychiatric illnesses: Are we inching
closer? Indian J Med Microbiol. 27:191–201. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang L, Xu MM, Zeng L, Liu S, Liu X, Wang
X, Li D, Huang RZ, Zhao LB, Zhan QL, et al: Evidence for Borna
disease virus infection in neuropsychiatric patients in three
western China provinces. Eur J Clin Microbiol Infect Dis.
33:621–627. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liu X, Bode L, Zhang L, Wang X, Liu S,
Zhang L, Huang R, Wang M, Yang L, Chen S, et al: Health care
professionals at risk of infection with Borna disease
virus-evidence from a large hospital in China (Chongqing). Virol J.
12:392015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hornig M, Briese T, Licinio J, Khabbaz RF,
Altshuler LL, Potkin SG, Schwemmle M, Siemetzki U, Mintz J,
Honkavuori K, et al: Absence of evidence for bornavirus infection
in schizophrenia, bipolar disorder and major depressive disorder.
Mol Psychiatry. 17:486–493. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Evengård B, Briese T, Lindh G, Lee S and
Lipkin WI: Absence of evidence of Borna disease virus infection in
Swedish patients with Chronic Fatigue Syndrome. J Neurovirol.
5:495–499. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wolff T, Heins G, Pauli G, Burger R and
Kurth R: Failure to detect Borna disease virus antigen and RNA in
human blood. J Clin Virol. 36:309–311. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Scordel C, Huttin A, Cochet-Bernoin M,
Szelechowski M, Poulet A, Richardson J, Benchoua A, Gonzalez-Dunia
D, Eloit M and Coulpier M: Borna disease virus phosphoprotein
impairs the developmental program controlling neurogenesis and
reduces human GABAergic neurogenesis. PLoS Pathog. 11:e10048592015.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hans A, Syan S, Crosio C, Sassone-Corsi P,
Brahic M and Gonzalez-Dunia D: Borna disease virus persistent
infection activates mitogen-activated protein kinase and blocks
neuronal differentiation of PC12 cells. J Biol Chem. 276:7258–7265.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hans A, Bajramovic JJ, Syan S, Perret E,
Dunia I, Brahic M and Gonzalez-Dunia D: Persistent, noncytolytic
infection of neurons by Borna disease virus interferes with ERK 1/2
signaling and abrogates BDNF-induced synaptogenesis. FASEB J.
18:863–865. 2004.PubMed/NCBI
|
|
21
|
Kamitani W, Ono E, Yoshino S, Kobayashi T,
Taharaguchi S, Lee BJ, Yamashita M, Kobayashi T, Okamoto M,
Taniyama H, et al: Glial expression of Borna disease virus
phosphoprotein induces behavioral and neurological abnormalities in
transgenic mice. Proc Natl Acad Sci USA. 100:8969–8974. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Mollaie HR, Monavari SH, Arabzadeh SA,
Shamsi-Shahrabadi M, Fazlalipour M and Afshar RM: RNAi and miRNA in
viral infections and cancers. Asian Pac J Cancer Prev.
14:7045–7056. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Krol J, Loedige I and Filipowicz W: The
widespread regulation of microRNA biogenesis, function and decay.
Nat Rev Genet. 11:597–610. 2010.PubMed/NCBI
|
|
26
|
Alvarez-Garcia I and Miska EA: MicroRNA
functions in animal development and human disease. Development.
132:4653–4662. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Liu X, Zhang L, Cheng K, Wang X, Ren G and
Xie P: Identification of suitable plasma-based reference genes for
miRNAome analysis of major depressive disorder. J Affect Disord.
163:133–139. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Fagan AM and Perrin RJ: Upcoming candidate
cerebrospinal fluid biomarkers of Alzheimer's disease. Biomark Med.
6:455–476. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Di Y, Lei Y, Yu F, Changfeng F, Song W and
Xuming M: MicroRNAs expression and function in cerebral ischemia
reperfusion injury. J Mol Neurosci. 53:242–250. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lages E, Ipas H, Guttin A, Nesr H, Berger
F and Issartel JP: MicroRNAs: Molecular features and role in
cancer. Front Biosci (Landmark Ed). 17:2508–2540. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Bode L, Dürrwald R, Rantam FA, Ferszt R
and Ludwig H: First isolates of infectious human Borna disease
virus from patients with mood disorders. Mol Psychiatry. 1:200–212.
1996.PubMed/NCBI
|
|
32
|
Huang R, Gao H, Zhang L, Jia J, Liu X,
Zheng P, Ma L, Li W, Deng J, Wang X, et al: Borna disease virus
infection perturbs energy metabolites and amino acids in cultured
human oligodendroglia cells. PLoS One. 7:e446652012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Liu X, Zhao L, Yang Y, Bode L, Huang H,
Liu C, Huang R, Zhang L, Wang X, Zhang L, et al: Human borna
disease virus infection impacts host proteome and histone lysine
acetylation in human oligodendroglia cells. Virology 464–465.
196–205. 2014. View Article : Google Scholar
|
|
34
|
Zhang L, Wang X, Zhan Q, Wang Z, Xu M, Zhu
D, He F, Liu X, Huang R, Li D, et al: Evidence for natural Borna
disease virus infection in healthy domestic animals in three areas
of western China. Arch Virol. 159:1941–1949. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhang L, Liu S, Zhang L, You H, Huang R,
Sun L, He P, Chen S, Zhang H and Xie P: Real-time qPCR identifies
suitable reference genes for Borna disease virus-infected rat
cortical neurons. Int J Mol Sci. 15:21825–21839. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhang L, Wang X, Zhan Q, Wang Z, Xu M, Zhu
D, He F, Liu X, Huang R, Li D, Lei Y and Xie P: Evidence for
natural Borna disease virus infection in healthy domestic animals
in three areas of western China. Arch Virol. 159:1941–1949. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Perkins JR, Dawes JM, McMahon SB, Bennett
DL, Orengo C and Kohl M: ReadqPCR and NormqPCR: R packages for the
reading, quality checking and normalisation of RT-qPCR
quantification cycle (Cq) data. BMC genomics. 13:2962012.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Li D, Lei Y, Deng J, Zhou C, Zhang Y, Li
W, Huang H, Cheng S, Zhang H, Zhang L, et al: Human but not
laboratory borna disease virus inhibits proliferation and induces
apoptosis in human oligodendrocytes in vitro. PLoS One.
8:e666232013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Liu X, Yang Y, Zhao M, Bode L, Zhang L,
Pan J, Lv L, Zhan Y, Liu S, Zhang L, et al: Proteomics reveal
energy metabolism and mitogen-activated protein kinase signal
transduction perturbation in human Borna disease virus
Hu-H1-infected oligodendroglial cells. Neuroscience. 268:284–296.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Lei Y, Li D, Deng J, et al: Metabolomic
profiling of three brain regions from a postnatal infected Borna
disease virus Hu-H1 rat model. Metabolomics. 10:484–495. 2014.
View Article : Google Scholar
|
|
41
|
Strickland ER, Hook MA, Balaraman S, Huie
JR, Grau JW and Miranda RC: MicroRNA dysregulation following spinal
cord contusion: Implications for neural plasticity and repair.
Neuroscience. 186:146–160. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zhao M, Sun L, Chen S, Li D, Zhang L, He
P, Liu X, Zhang L, Zhang H, Yang D, et al: Borna disease virus
infection impacts microRNAs associated with nervous system
development, cell differentiation, proliferation and apoptosis in
the hippocampi of neonatal rats. Mol Med Rep. 12:3697–3703.
2015.PubMed/NCBI
|
|
43
|
Liu Z, Liu Y, Li L, Xu Z, Bi B, Wang Y and
Li JY: MiR-7-5p is frequently downregulated in glioblastoma
microvasculature and inhibits vascular endothelial cell
proliferation by targeting RAF1. Tumour Biol. 35:10177–10184. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Fang Y, Xue JL, Shen Q, Chen J and Tian L:
MicroRNA-7 inhibits tumor growth and metastasis by targeting the
phosphoinositide 3-kinase/Akt pathway in hepatocellular carcinoma.
Hepatology. 55:1852–1862. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Giles KM, Brown RA, Epis MR, Kalinowski FC
and Leedman PJ: miRNA-7-5p inhibits melanoma cell migration and
invasion. Biochem Biophys Res Commun. 430:706–710. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Shi Y, Luo X, Li P, Tan J, Wang X, Xiang T
and Ren G: miR-7-5p suppresses cell proliferation and induces
apoptosis of breast cancer cells mainly by targeting REGγ. Cancer
Lett. 358:27–36. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Saydam O, Senol O, Würdinger T, Mizrak A,
Ozdener GB, Stemmer-Rachamimov AO, Yi M, Stephens RM, Krichevsky
AM, Saydam N, et al: miRNA-7 attenuation in Schwannoma tumors
stimulates growth by upregulating three oncogenic signaling
pathways. Cancer Res. 71:852–861. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Wu DG, Wang YY, Fan LG, Luo H, Han B, Sun
LH, Wang XF, Zhang JX, Cao L, Wang XR, et al: MicroRNA-7 regulates
glioblastoma cell invasion via targeting focal adhesion kinase
expression. Chin Med J (Engl). 124:2616–2621. 2011.PubMed/NCBI
|
|
49
|
Kong X, Li G, Yuan Y, He Y, Wu X, Zhang W,
Wu Z, Chen T, Wu W, Lobie PE and Zhu T: MicroRNA-7 inhibits
epithelial-to-mesenchymal transition and metastasis of breast
cancer cells via targeting FAK expression. PLoS One. 7:e415232012.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Zhao X, Dou W, He L, Liang S, Tie J, Liu
C, Li T, Lu Y, Mo P, Shi Y, et al: MicroRNA-7 functions as an
anti-metastatic microRNA in gastric cancer by targeting
insulin-like growth factor-1 receptor. Oncogene. 32:1363–1372.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Wu K, Hu G, He X, Zhou P, Li J, He B and
Sun W: MicroRNA-424-5p suppresses the expression of SOCS6 in
pancreatic cancer. Pathol Oncol Res. 19:739–748. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Zhang Y, Li T, Guo P, Kang J, Wei Q, Jia
X, Zhao W, Huai W, Qiu Y, Sun L and Han L: MiR-424-5p reversed
epithelial-mesenchymal transition of anchorage-independent HCC
cells by directly targeting ICAT and suppressed HCC progression.
Sci Rep. 4:62482014. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Bai Y, Liao H, Liu T, Zeng X, Xiao F, Luo
L, Guo H and Guo L: MiR-296-3p regulates cell growth and multi-drug
resistance of human glioblastoma by targeting ether-à-go-go (EAG1).
Eur J Cancer. 49:710–724. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Kolodziejek J, Dürrwald R, Herzog S,
Ehrensperger F, Lussy H and Nowotny N: Genetic clustering of Borna
disease virus natural animal isolates, laboratory and vaccine
strains strongly reflects their regional geographical origin. J Gen
Virol. 86:385–398. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Zhai A, Qian J, Kao W, Li A, Li Y, He J,
Zhang Q, Song W, Fu Y, Wu J, et al: Borna disease virus encoded
phosphoprotein inhibits host innate immunity by regulating miR-155.
Antiviral Res. 98:66–75. 2013. View Article : Google Scholar : PubMed/NCBI
|