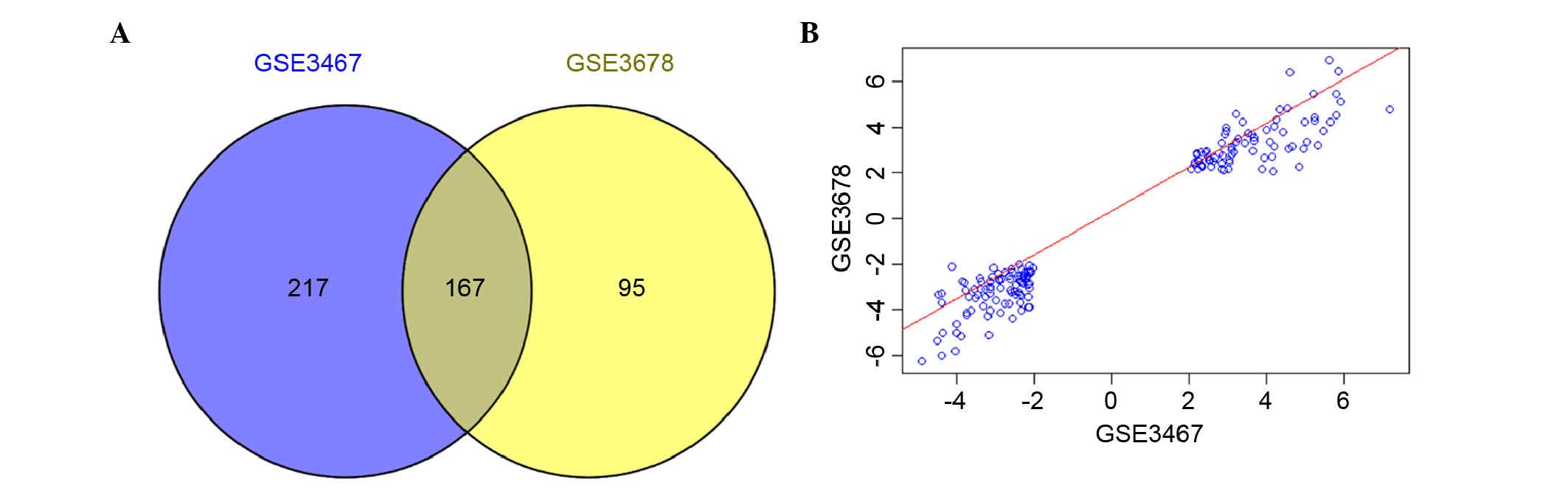
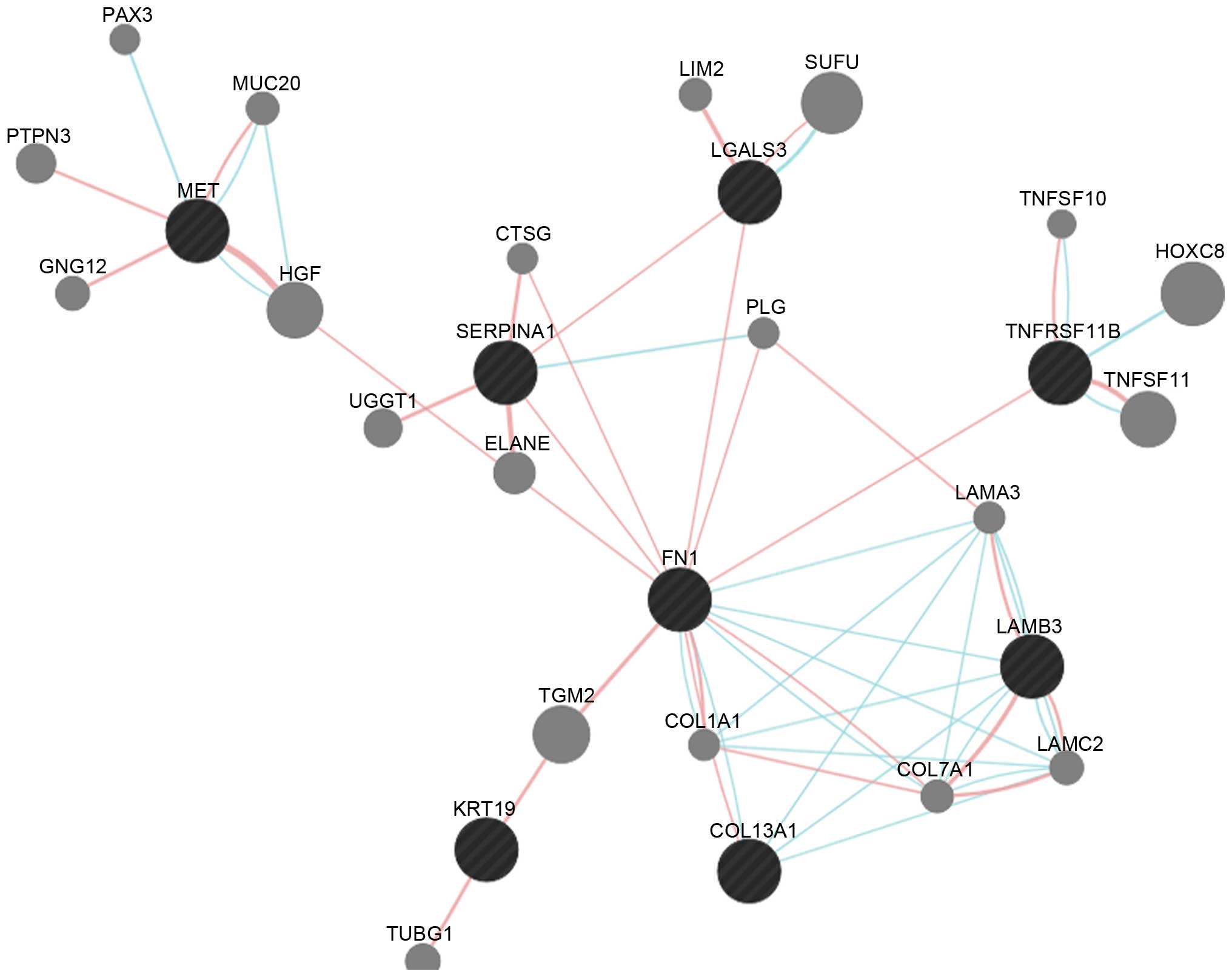
|
1
|
Nguyen QT, Lee EJ, Huang MG, Park YI,
Khullar A and Plodkowski RA: Diagnosis and treatment of patients
with thyroid cancer. Am Health Drug Benefits. 8:30–40.
2015.PubMed/NCBI
|
|
2
|
Siegel R, Ma J, Zou Z and Jemal A: Cancer
statistics, 2014. CA Cancer J Clin. 64:9–29. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
National Cancer Institute, . SEER stat
fact sheets: Thyroid cancer. http://seer.cancer.gov/statfacts/html/thyro.htmlAccessed:
May 20, 2015.
|
|
4
|
Schneider DF and Chen H: New developments
in the diagnosis and treatment of thyroid cancer. CA Cancer J Clin.
63:374–394. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yu XM, Wan Y, Sippel RS and Chen H: Should
all papillary thyroid microcarcinomas be aggressively treated? An
analysis of 18,445 cases. Ann Surg. 254:653–660. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Mazeh H and Chen H: Advances in surgical
therapy for thyroid cancer. Nat Rev Endocrinol. 7:581–588. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Cohen Y, Xing M, Mambo E, Guo Z, Wu G,
Trink B, Beller U, Westra WH, Ladenson PW and Sidransky D: BRAF
mutation in papillary thyroid carcinoma. J Natl Cancer Inst.
95:625–627. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Soares P, Trovisco V, Rocha AS, Lima J,
Castro P, Preto A, Máximo V, Botelho T, Seruca R and
Sobrinho-Simões M: BRAF mutations and RET/PTC rearrangements are
alternative events in the etiopathogenesis of PTC. Oncogene.
22:4578–4580. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Fukushima T, Suzuki S, Mashiko M, Ohtake
T, Endo Y, Takebayashi Y, Sekikawa K, Hagiwara K and Takenoshita S:
BRAF mutations in papillary carcinomas of the thyroid. Oncogene.
22:6455–6457. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhang H, Teng X and Liu Z, Zhang L and Liu
Z: Gene expression profile analyze the molecular mechanism of CXCR7
regulating papillary thyroid carcinoma growth and metastasis. J Exp
Clin Cancer Res. 34:162015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yang Z, Fan Y, Deng Z, Wu B and Zheng Q:
Amyloid precursor protein as a potential marker of malignancy and
prognosis in papillary thyroid carcinoma. Oncol Lett. 3:1227–1230.
2012.PubMed/NCBI
|
|
12
|
He H, Jazdzewski K, Li W, Liyanarachchi S,
Nagy R, Volinia S, Calin GA, Liu CG, Franssila K, Suster S, et al:
The role of microRNA genes in papillary thyroid carcinoma. Proc
Natl Acad Sci USA. 102:19075–19080. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhu X, Yao J and Tian W: Microarray
technology to investigate genes associated with papillary thyroid
carcinoma. Mol Med Rep. 11:3729–3733. 2015.PubMed/NCBI
|
|
14
|
Barrett T, Troup DB, Wilhite SE, Ledoux P,
Rudnev D, Evangelista C, Kim IF, Soboleva A, Tomashevsky M and
Edgar R: NCBI GEO: Mining tens of millions of expression
profiles-database and tools update. Nucleic Acids Res. 35:(Database
issue). D760–D765. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gentleman RC, Carey VJ, Bates DM, Bolstad
B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al:
Bioconductor: Open software development for computational biology
and bioinformatics. Genome Biol. 5:R802004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Smyth GK: Limma: linear models for
microarray dataBioinformatics and computational biology solutions
using R and Bioconductor. Gentleman R, Varey VJ, Huber W, Irizarry
RA and Dudoit S: 1st. Springer-Verlag; New York, NY: pp. 397–420.
2005, View Article : Google Scholar
|
|
17
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Harris MA, Clark J, Ireland A, Lomax J,
Ashburner M, Foulger R, Eilbeck K, Lewis S, Marshall B, Mungall C,
et al: The Gene Ontology (GO) database and informatics resource.
Nucleic Acids Res. 32:(Database issue). D258–D261. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kanehisa M and Goto S: KEGG: Kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Warde-Farley D, Donaldson SL, Comes O,
Zuberi K, Badrawi R, Chao P, Franz M, Grouios C, Kazi F, Lopes CT,
et al: The GeneMANIA prediction server: Biological network
integration for gene prioritization and predicting gene function.
Nucleic acids Res. 38:(Web Server issue). W214–W220. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Bader GD and Hogue CW: An automated method
for finding molecular complexes in large protein interaction
networks. BMC Bioinformatics. 4:22003. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wishart DS, Knox C, Guo AC, Shrivastava S,
Hassanali M, Stothard P, Chang Z and Woolsey J: DrugBank: A
comprehensive resource for in silico drug discovery and
exploration. Nucleic Acids Res. 34:(Database issue). D668–D672.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Labbé CM, Rey J, Lagorce D, Vavruša M,
Becot J, Sperandio O, Villoutreix BO, Tufféry P and Miteva MA:
MTiOpenScreen: A web server for structure-based virtual screening.
Nucleic Acids Res. 43:W448–W454. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lipinski CA, Lombardo F, Dominy BW and
Feeney PJ: Experimental and computational approaches to estimate
solubility and permeability in drug discovery and development
settings. Adv Drug Deliv Rev. 46:3–26. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Collen D and Lijnen HR: Tissue-type
plasminogen activator: A historical perspective and personal
account. J Thromb Haemostasis. 2:541–546. 2004. View Article : Google Scholar
|
|
26
|
Komminoth P: The RET proto-oncogene in
medullary and papillary thyroid carcinoma. Molecular features,
pathophysiology and clinical implications. Virchows Arch. 431:1–9.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Eberhardt NL, Grebe SK, McIver B and Reddi
HV: The role of the PAX8/PPARgamma fusion oncogene in the
pathogenesis of follicular thyroid cancer. Mol Cell Endocrinol.
321:50–56. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Tanaka T, Umeki K, Yamamoto I, Sugiyama S,
Noguchi S and Ohtaki S: Immunohistochemical loss of thyroid
peroxidase in papillary thyroid carcinoma: Strong suppression of
peroxidase gene expression. J Pathol. 179:89–94. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ruf J and Carayon P: Structural and
functional aspects of thyroid peroxidase. Arch Biochem Biophys.
445:269–277. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Buitrago D, Keutgen X, Crowley M, Filicori
F, Aldailami H, Hoda R, Liu YF, Hoda RS, Scognamiglio T, Jin M, et
al: Intercellular adhesion molecule-1 (ICAM-1) is upregulated in
aggressive papillary thyroid carcinoma. Ann Surg Oncol. 19:973–980.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Jarząb B, Wiench M, Fujarewicz K, Simek K,
Jarzab M, Oczko-Wojciechowska M, Wloch J, Czarniecka A, Chmielik E,
Lange D, et al: Gene expression profile of papillary thyroid
cancer: Sources of variability and diagnostic implications. Cancer
Res. 65:1587–1597. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Griffith OL, Melck A, Jones SJ and Wiseman
SM: Meta-analysis and meta-review of thyroid cancer gene expression
profiling studies identifies important diagnostic biomarkers. J
Clin Oncol. 24:5043–5051. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
da Silveira Mitteldorf CA, de
Sousa-Canavez JM, Leite KR, Massumoto C and Camara-Lopes LH: FN1,
GALE, MET, and QPCT overexpression in papillary thyroid carcinoma:
Molecular analysis using frozen tissue and routine fine-needle
aspiration biopsy samples. Diagn Cytopathol. 39:556–561. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Vasko V, Espinosa AV, Scouten W, He H,
Auer H, Liyanarachchi S, Larin A, Savchenko V, Francis GL, de la
Chapelle A, et al: Gene expression and functional evidence of
epithelial-to-mesenchymal transition in papillary thyroid carcinoma
invasion. Proc Natl Acad Sci USA. 104:2803–2808. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yang Z, Yuan Z, Fan Y, Deng X and Zheng Q:
Integrated analyses of microRNA and mRNA expression profiles in
aggressive papillary thyroid carcinoma. Mol Med Rep. 8:1353–1358.
2013.PubMed/NCBI
|
|
36
|
Wasenius VM, Hemmer S, Kettunen E,
Knuutila S, Franssila K and Joensuu H: Hepatocyte growth factor
receptor, matrix metalloproteinase-11, tissue inhibitor of
metalloproteinase-1, and fibronectin are up-regulated in papillary
thyroid carcinoma: A cDNA and tissue microarray study. Clin Cancer
Res. 9:68–75. 2003.PubMed/NCBI
|
|
37
|
Vierlinger K, Mansfeld MH, Koperek O,
Nöhammer C, Kaserer K and Leisch F: Identification of SERPINA1 as
single marker for papillary thyroid carcinoma through microarray
meta analysis and quantification of its discriminatory power in
independent validation. BMC Med Genomics. 4:302011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Lewy-Trenda I: Estrogen and progesterone
receptors in neoplastic and non-neoplastic thyroid lesions. Pol J
Pathol. 53:67–72. 2002.PubMed/NCBI
|
|
39
|
Kawabata W, Suzuki T, Moriya T, Fujimori
K, Naganuma H, Inoue S, Kinouchi Y, Kameyama K, Takami H,
Shimosegawa T and Sasano H: Estrogen receptors (alpha and beta) and
17beta-hydroxysteroid dehydrogenase type 1 and 2 in thyroid
disorders: Possible in situ estrogen synthesis and actions. Mod
Pathol. 16:437–444. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Derewenda ZS and Derewenda U: The
structure and function of platelet-activating factor
acetylhydrolases. Cell Mol Life Sci. 54:446–455. 1998. View Article : Google Scholar : PubMed/NCBI
|