Introduction
The mortality rate associated with colon cancer
ranks as the fourth highest of different cancer types worldwide
(1). According to the data from
Globocan, an international cancer research organization governed by
the World Health Organization, there were 694,000 cases of colon
cancer-associated mortality during 2012, and 52% of these were from
developing countries. Eastern Europe had the highest mortality rate
at 20.3% in males and 11.7% in females, while the Western Africa
had the lowest mortality rate at 3.5% in males and 3.0% in females.
There are 253,000 recently diagnosed cases in China, predominantly
in 50–55 year olds. The morbidity is increasing in males, and is
marginally reducing in females in China (2).
TFAM, which is a member of the high mobility group
(HMG) box protein family, participates in the process of
mitochondrial DNA (mtDNA) replication and transcription (3–5). In
numerous types of cancer cells, HMG proteins are upregulated
(6). In addition, mitochondria are
critical for cancer cell involvement in the regulation of cancer
cell survival and growth. The Warburg effect is essential for the
survival and proliferation of cancer cells by mitochondrial
uncoupling regulating the metabolic shift to aerobic glycolysis
(7). It has been reported that the
high expression of TFAM is associated with a poor prognosis in
multiple malignant tumors, including ovarian cancer, endometrial
carcinoma and colon cancer (8,9). It
has been reported that the TFAM protein multimerizes and binds to
mtDNA which regulates the expression of nuclear genes, suggesting
that the TFAM levels may be increased in cancer cells and be
associated with malignant progression and proliferative activity
(10–13). However, the roles of TFAM remain to
be fully elucidated in colon cancer cells.
miRNAs, which are small, endogenous and non-coding
RNAs, can interact with target sites in the 3′-untranslated regions
(UTRs) of mRNA and then inhibit the expression of targeted genes
(14,15). Those targeted genes are involved in
the regulation of multiple biological processes, including the
pathogenesis of a variety of types of human cancer (16–22).
miR-590-3p has been reported to be involved in mediating the
expression of autoimmune genes and neuronal death (23,24).
However, whether or not miR-590-3p is associated with colon cancer
remains unclear. Thus, the present study aimed to elucidate the
roles of TFAM and miR-590-3p and their association in colon cancer
cells.
Materials and methods
Human tissue specimens
The present study was approved by the ethics
committee of the First Affiliated Hospital of Sun Yat-Sen
University (Guangzhou, China). There were 30 cases of colectomy for
colon cancer performed by the First Affiliated Hospital of Sun
Yat-Sen University between March and April 2013. Samples were
collected from the specimens at the mucosa that were 10 cm adjacent
to the colon cancer (the control group), and from the cancer tissue
itself (the experiment group). Patients provided written informed
consent.
Patient selection criteria: i) Primary colon cancer
diagnosed by histopathology; ii) underwent colon cancer resection
and lymph gland cleaning; iii) a minimum of 12 lymph glands
undergoing biopsy subsequent to surgery; iv) detailed pathological
follow-up records.
Patient rejection criteria: i) History of an
additional primary cancer; ii) distant metastasis (peritoneum,
liver, brain, etc.); iii) underwent neoadjuvant chemotherapy and
other etiotropic therapy; iv) succumbed to disease.
A total of 30 patients were selected, with an
average age of 59 years old and including 18 males (60%) and 12
females (40%). The cancer locations were identified, with 15 cases
(50%) at the sigmoid colon, 7 cases (23.3%) at the descending colon
and 8 cases (26.7%) at the ascending colon. According to the TNM
classification in the 3rd edition of NCCN 2014 (25), there were 28 phase III patients
(93.3%) and 2 phase II patients (6.7%).
Specimen collection involved obtaining ~5×5 mm
samples from the cancer tissues and from mucosa that was 10 cm
adjacent to the cancer (normal colon tissue) within 20 min of
removal from the body. The specimens were placed into two tubes and
stored in liquid nitrogen; one to be used for protein extraction
and the other for RNA extraction.
Cell culture
Normal human colon cells were purchased from the
American Type Culture Collection (CCD-18Co; ATCC®
CRL-1459™; Manassas, VA, USA). SW480 cells (Institute of Cell
Biology, Chinese Academy of Sciences, Shanghai, China) was cultured
in RPMI-1640 medium with 10% fetal bovine serum (FBS) and 1%
penicillin/streptomycin (Xinxianfeng Pharmaceutical Industry
Limited Company, Shanghai, China) at 37°C, with 5% CO2. The
lentivirus, lentiviral vector and Dual-Luciferase Reporter Assays
were purchased from GeneCopoeia, Inc. (Guangzhou, China).
miRNA prediction
Subsequent to inputting TFAM to miRWalk 2.0
(http://zmf.umm.uni-heidelberg.de/apps/zmf/mirwalk2/)
as target gene, matching miRNA results were indicated by high
scores in Microt4, miRanda, miRBridge, miRDB, miRMap, miRNAMap,
PicTar2, PITA, RNA22, RNAhybrid and TargetScan. The target miRNAs
were verified in TargetScan and miRanda.
RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
RNA was extracted from the normal colon tissues (30
specimens), colon cancer tissues (30 specimens), normal colon cells
and SW480 cells using TRIzol reagent (Invitrogen; Thermo Fisher
Scientific, Inc., Waltham, MA, USA), according to the
manufacturer's instructions. The RNA integrity was then assessed. A
TaqMan RT-qPCR miRNA assay (Applied Biosystems; Thermo Fisher
Scientific, Inc.) was performed to detect the expression levels of
mature miR-590-3p. The relative expression of mature miR-590-3p
levels normalized to U6 endogenous control was determined using the
2−ΔΔCq method (26).
Each measurement was performed in triplicate. In order to detect
the target genes (mRNA expression), 1 µg total RNA was reverse
transcribed to cDNA using SuperScript™ III First-Strand Synthesis
Super Mix (Invitrogen; Thermo Fisher Scientific, Inc.). SYBR green
qPCR was performed using the Bio-Rad iQ5 PCR detection system
(Bio-Rad Laboratories, Inc., Hercules, CA, USA) with the following
gene-specific primers: miR-590-3p, forward
5′-AAAGATTCCAAGAAGCTAAGGGTG-3′, reverse
5′-CCTAACTGGTTTCCTGTGCCTA-3′; and U6, forward
5′-TGCGGGTGCTCCGCTTCGGCAGC-3′ and reverse 5′-CAGTGCAGGGTCCGAGGT-3′.
PCR cycling conditions were as follows: Initial cycle at 95°C for 3
min, followed by pre-denaturation at 95°C for 15 sec, denaturation
at 95°C for 5 sec and extension at 60°C for 30 sec, for 45 cycles,
and a final extension step at 70°C for 30 min.
Correlation analysis
Correlation analysis was conducted for the
expression of miR-590-3p mRNA (normalized to U6) and the expression
of TFAM mRNA (normalized to β-actin) in the same specimen.
Dual luciferase reporter assays
To generate the reporter vectors bearing
miRNA-binding sites, a normal and mutated 3′-UTR of TFAM was
sub-cloned using PCR-based methods (27). The constructs were inserted into
the multiple cloning sites downstream of the luciferase gene in the
psiCHECK-2 luciferase miRNA expression reporter vector. For the
luciferase assay, 1×105 cells were cultured to 70–80%
confluence in 24-well plates and co-transfected with
psiCHECK-2-TFAM-3′-UTR or psiCHECK2-mut-TFAM-3′-UTR vector plus 50
nM miR-590-3p or 100 mM miR-590-3p inhibitor using Lipofectamine
2000 (Invitrogen; Thermo Fisher Scientific, Inc.), according to the
manufacturer's instructions. The cells were incubated with
transfection reagent/DNA complex for 5 h at 37°C and refreshed with
fresh medium containing 10% FBS. At 48 h post-transfection, firefly
and Renilla luciferase activities were evaluated using the
Dual-Luciferase Reporter Assay system and the Renilla
luciferase activity was normalized to firefly luciferase
activity.
Western blotting
The expression of TFAM in SW480 cells was determined
subsequent to transfection by miR-590-3p lentivirus or
anti-miR-590-3p lentivirus by western blotting. Following 48
h-transfection and 24 h-cell culture, the cells were solubilized in
cold RIPA lysis buffer and then separated with 10% SDS-PAGE.
Following this, the proteins were transferred onto PVDF membranes.
The membranes were blocked in 5% skimmed dried milk in
phosphate-buffered saline and then incubated overnight with primary
antibodies for TFAM (1:500; cat. no. PB9447; Boster Biological
Technology, Pleasanton, CA, China), and β-actin (1:500; cat. no.
PA1872; Boster Biological Technology). Subsequent to incubation
with goat polyclonal anti-rabbit secondary antibodies (1:4,000;
cat. no. A-11034; Invitrogen; Thermo Fisher Scientific, Inc.), the
immune complexes were detected using the enhanced chemiluminescence
method. The blots were visualized by autoradiography using
preflashed Kodak XAR film (Kodak Japan Ltd., Tokyo, Japan). The
results were analyzed with Gel-Pro Analyzer, version 4.0 (Silk
Scientific, Inc., Orem, UT USA) subsequent to visualization.
MTT assays
Subsequent to transfection, the miR-SCR-SW480,
miR-590-3p-SW480 and anti-miR-590-3p-SW480 cell groups were
cultured at a density of 5,000 cells/well. A total of 10 µl MTT (5
mg/ml; Promega Corporation) was added to the medium at 0, 24, 48,
72, 96 and 120 h. The cells were then incubated for 4 h at 37°C.
The medium was removed and 100 µl dimethyl sulfoxide was added into
each well. The plate was gently rotated on an orbital shaker for 15
min to completely dissolve the precipitation. The absorbance was
detected at 490 nm with a microplate reader (Bio-Rad Laboratories,
Inc.).
Statistical analysis
All experiments were performed three times in
triplicate. The data were analyzed with Student's two-tailed
t-test. All statistical analyses were performed using SPSS
software, version 17.0 (SPSS, Inc., Chicago, IL, USA). P<0.05
was considered to indicate a statistically significant
difference.
Results
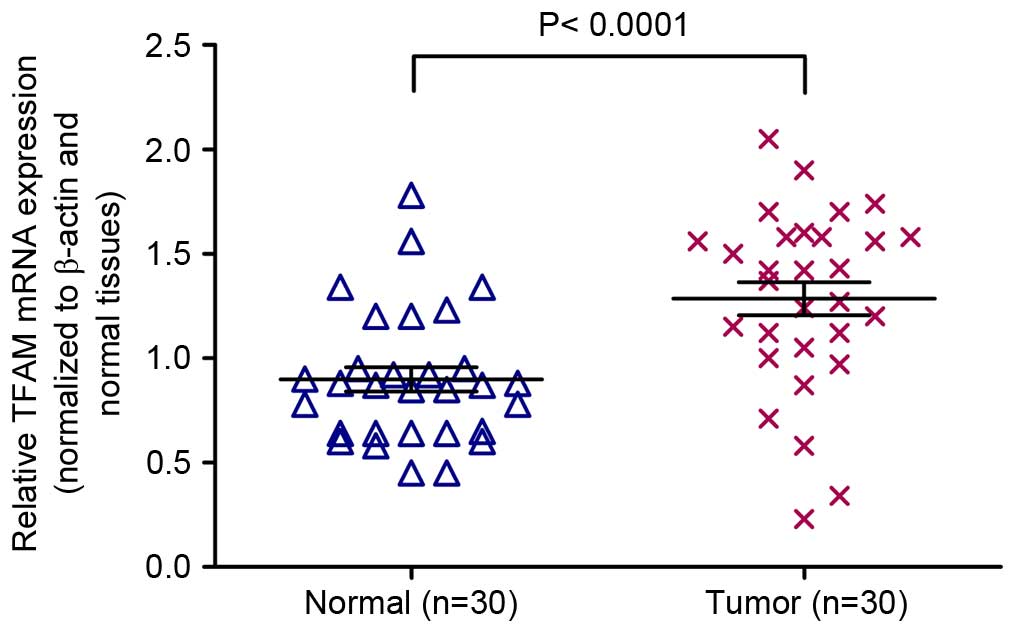
mRNA expression of TFAM is increased
in colon cancer tissues vs. adjacent normal colon tissues
mRNA expression of TFAM in tumor tissue samples and
adjacent normal tissues was examined using the RT-qPCR assay. The
average expression levels of TFAM were significantly increased in
colon cancer tissues compared with adjacent normal colon tissues
(Fig. 1; P<0.0001).
Correlation analysis between TFAM and
miR-590-3p
miR-590-3p exhibited a high prediction score (the
highest score is 9) in miRWalk 2.0 (Fig. 2A), identifying TFAM as the target
gene. The 3′-UTR of miR-590-3p was correlated with TFAM in miRanda
(Fig. 2B) and TargetScan (Fig. 2C). The results of the luciferase
assay for miR-590-3p are presented in Fig. 2D and the data indicated that the
Renilla/firefly value of luciferase was significantly lower
in the miR-590-3p treatment cells following transfection with the
3′-UTR of the TFAM gene (Psi-wtTFAM; P=0.005), while the
Renilla/firefly value of luciferase exhibited no differences
following transfection with the mutated 3′-UTR of TFAM
(Psi-mutTFAM) compared with the control (Fig. 2D; P=0.7870). These data suggested
the 3′-UTR of TFAM is the direct target of miR-590-3p. The
expression of miR-590-3p (normalized to U6) in normal colon and
colon cancer tissues exhibited positive correlation with that of
TFAM (normalized to β-actin) in the same specimens, correlation
coefficent r=0.9456, P<0.0001 (Fig.
2E).
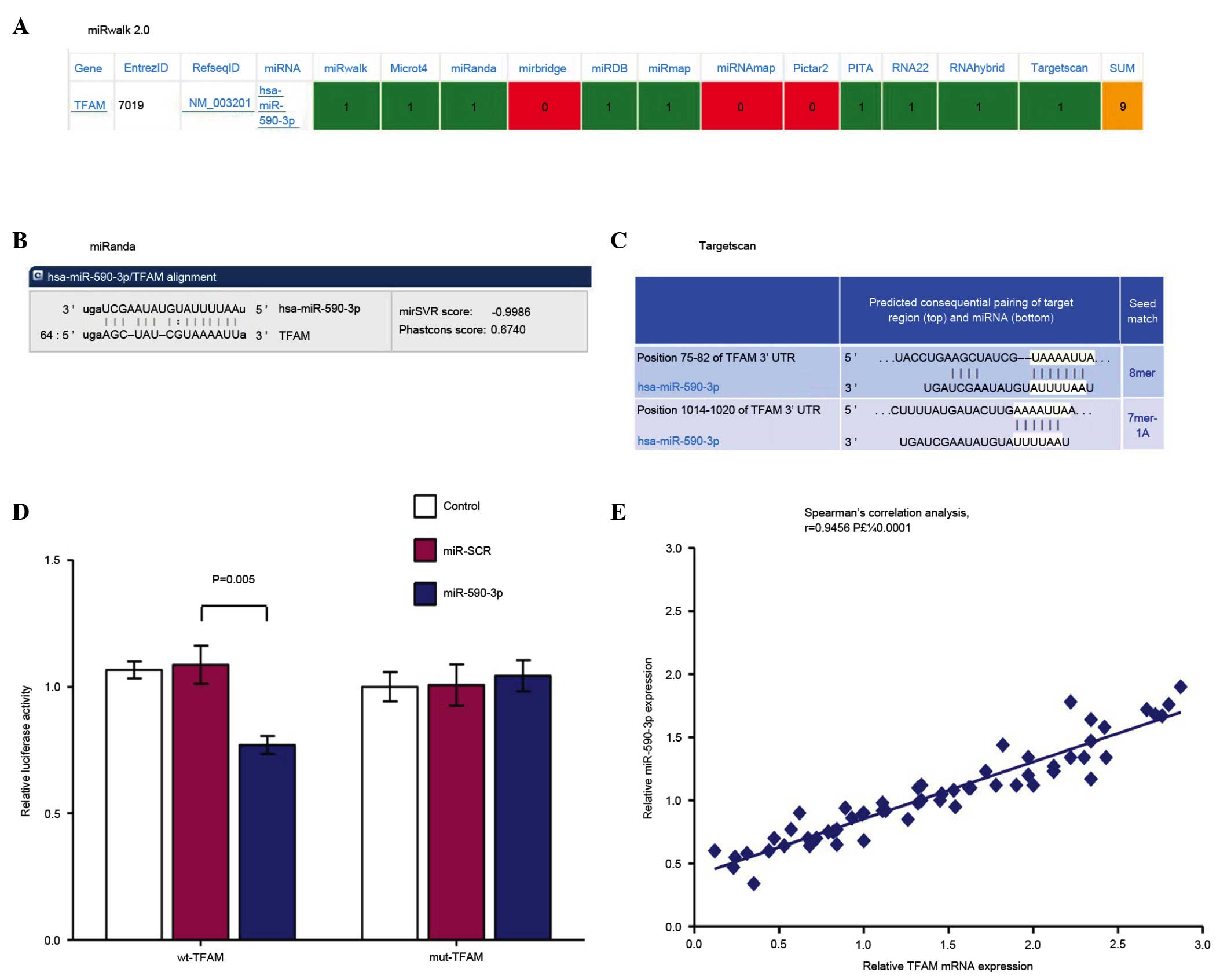 | Figure 2.Correlation analysis between TFAM and
miR-590-3p. (A) miR-590-3p demonstrated a high prediction score
(the highest score is 9) in miRWalk 2.0, matching TFAM as the
target gene. The 3′-UTR of miR-590-3p matched with TFAM in (B)
miRanda and (C) TargetScan. (D) Luciferase assay results for
miR-590-3p; the data indicated that the renilla/firefly value of
luciferase was significantly lower in miR-590-3p-treatment cells
following transfection with the 3′-UTR of the TFAM gene
(Psi-wtTFAM, P=0.005), while the renilla/firefly value of
luciferase exhibited no difference following transfection with the
mutated 3′-UTR of TFAM (Psi-mutTFAM) compared with the control
(P=0.7870). These data suggested the 3′-UTR of TFAM is a direct
target of miR-590-3p. (E) The expression of miR-590-3p (normalized
to U6) in normal colon and colon cancer tissues were observed to
exhibit a positive correlation with that of TFAM (normalized to
β-actin) in the same specimen, correlation coefficent r=0.9456,
P<0.0001. TFAM, mitochondrial transcription factor A; miR,
microRNA; UTR, untranslated region; control, SW480 cells; miR-SCR,
SW480 cells transfected with miR-SCR lentivirus; miR-590-3p, SW480
cells transfected with miR-590-3p lentivirus. |
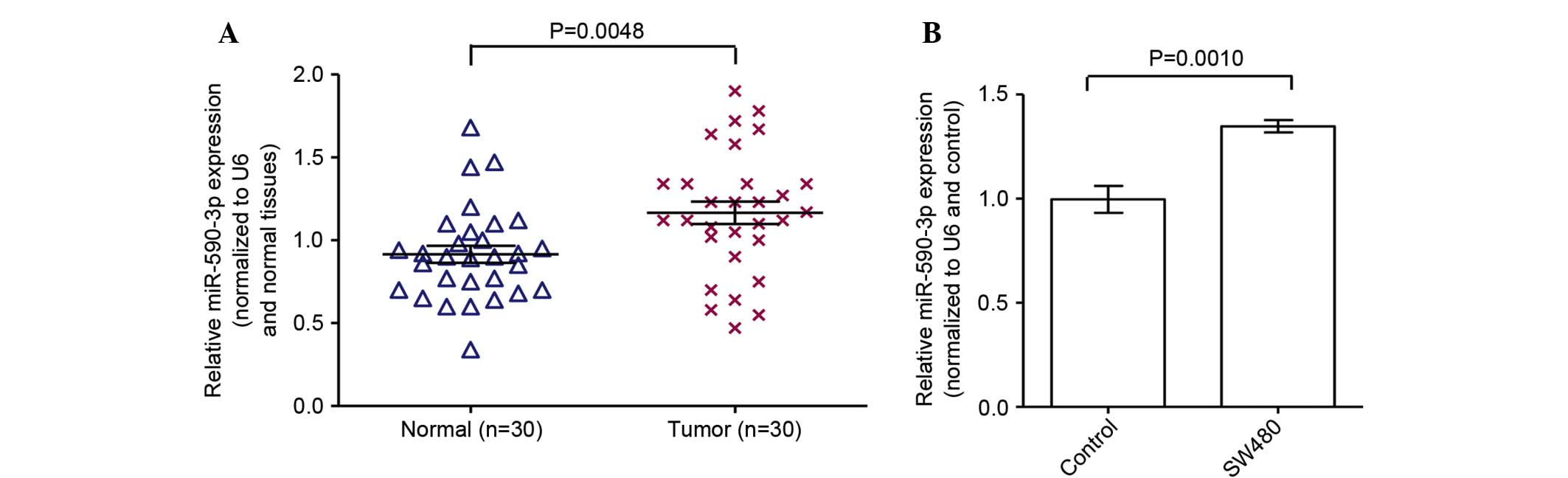
mRNA expression of miR-590-3p is
increased in colon cancer tissues and cells
mRNA expression of miR-590-3p in tumor tissue
samples and adjacent normal tissues was examined through the
RT-qPCR assay. The expression of miR-590-3p in the colon cancer
tissues was significantly increased compared with adjacent normal
colon tissues (Fig. 3A;
P<0.05). In addition, the mRNA expression of miR-590-3p in SW480
cells was significantly increased compared with the normal colon
cells (Fig. 3A; P<0.05).
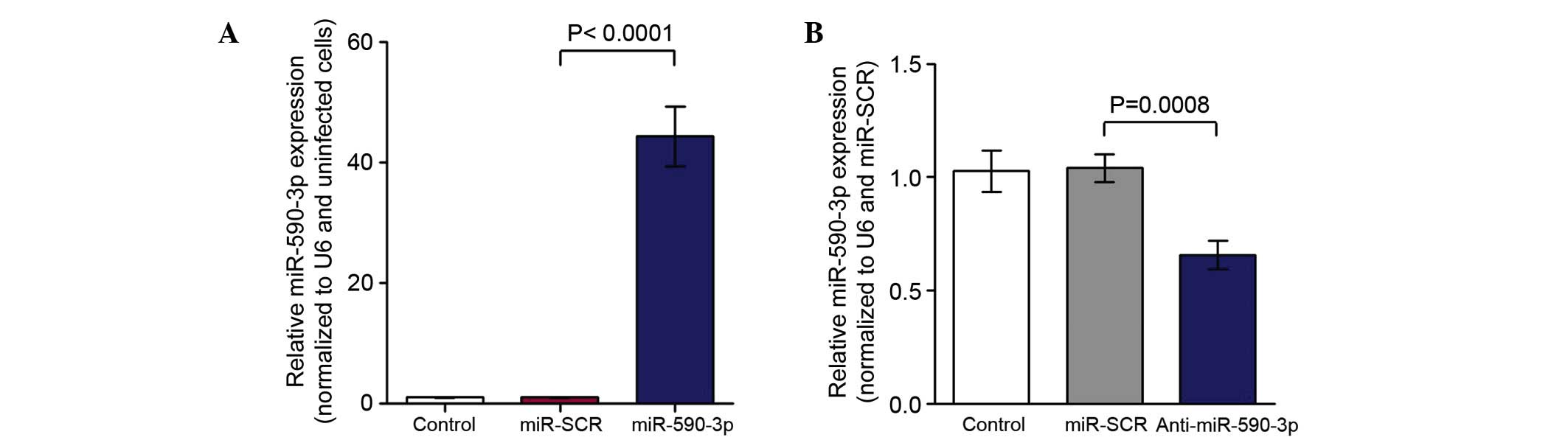
The miR-590-3p inhibitor effectively
suppressed miR-590-3p expression
Based on the fact that the expression level of
miR-590-3p is upregulated in colon cancer tissues and cells, the
miR-590-3p inhibitor was transfected into SW480 cells. Subequently,
cells were harvested for RT-qPCR. The results indicated that the
mRNA expression of miR-590-3p was increased in SW480 cells
transfected with miR-590-3p lentivirus compared with the control
(Fig. 4A; P<0.0001). The
expression of miR-590-3p mRNA was reduced in the SW480 cells
transfected with the anti-miR-590-3p lentivirus compared with that
of the control (Fig. 4B;
P=0.0008).
Inhibition of miR-590-3p suppressed
TFAM protein expression
To demonstrate the role of miR-590-3p in regulating
TFAM protein expression in SW480 cells. Western blotting was
conducted subsequent to transfection with miR-590-3p inhibitor
(anti-miR-590-3p) or miR-SCR. The expression of TFAM was
significantly increased in the SW480 cells transfected with
miR-590-3p lentivirus when compared with that of the control
(Fig. 5A and B; P<0.0001). The
expression of TFAM was observed to be significantly reduced in the
SW480 cells transfected with the anti-miR-590-3p lentivirus
compared with that of the control (Fig. 5C and D; P=0.0002).
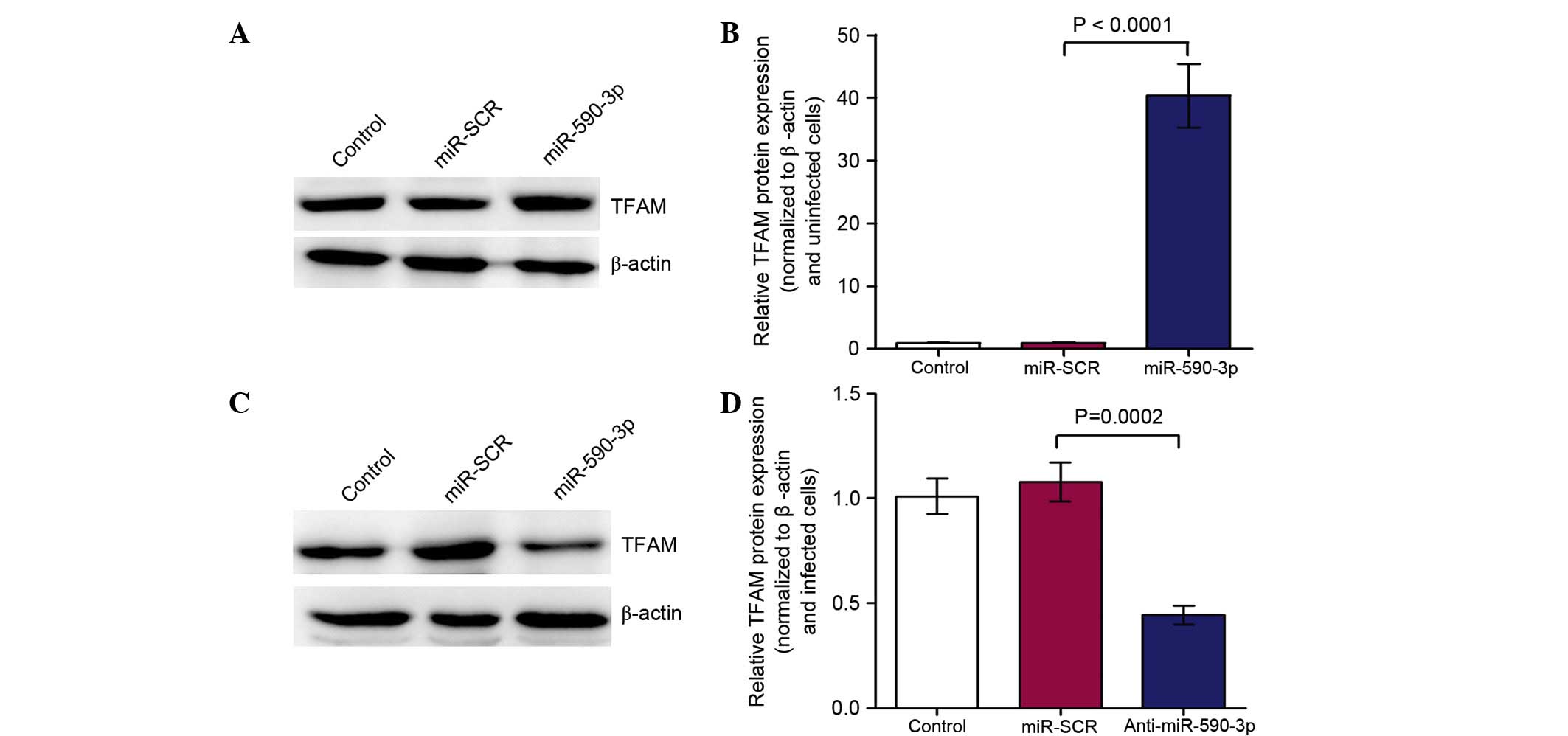 | Figure 5.Western blotting was conducted
following transfection with the miR-590-3p inhibitor
(anti-miR-590-3p) or miR-SCR. (A and B) TFAM expression was
significantly higher (P<0.0001) in SW480 cells transfected with
miR-590-3p lentivirus vs. the control (A, western blots; B,
quantification). (C and D) The expression of TFAM was significantly
(P=0.0002) lower in SW480 cells transfected with anti-miR-590-3p
lentivirus when compared with the control (C, western blots; D,
quantification). miR, microRNA; TFAM, mitochondrial transcription
factor A; control, SW480 cells; miR-SCR, SW480 cells transfected
with miR-SCR lentivirus; miR-590-3p, SW480 cells transfected with
miR-590-3p lentivirus; anti-miR-590-3p, SW480 cells transfected
with the anti-miR-590-3p lentivirus. |
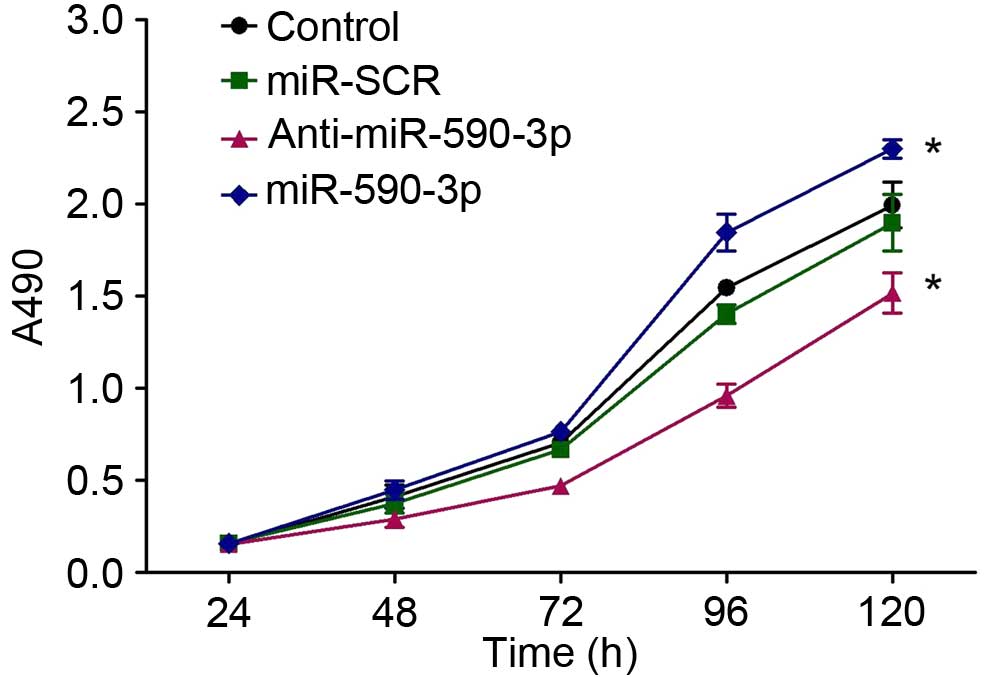
Inhibition of miR-590-3p suppressed
the proliferation of SW480 cells
In order to demonstrate the role of miR-590-3p in
the regulation of SW480 cell proliferation, MTT assays were
conducted subsequent to transfection with the miR-590-3p inhibitor
(anti-miR-590-3p) or miR-SCR. In the MTT assays, the proliferation
of the four groups were observed to have no significant differences
in the first 96 h following transfection. However, a growth
inhibition of SW480 cells transfected with anti-miR-590-3p compared
with controls (P<0.0001) was observed 120 h after transfection.
The proliferation of SW480 cells with miR-590-3p transfection was
greater than that of the controls (P<0.0001) while that between
SW480 and miR-SCR-SW480 exhibited no differences (P>0.05) 120 h
subsequent to transfection (Fig.
6).
Discussion
It has been reported that miRNAs serve significant
roles in tumorigenesis. Studies have demonstrated that certain
miRNAs are significantly deregulated in gastric cancer, bladder
cancer, endometrial carcinoma and may function as tumor regulators
(28–30). miRNAs represent a class of small
interfering RNAs that can silence genes. miRNAs hybridize to
partially complementary binding sites that are typically located in
the 3-UTR of target mRNAs and regulate their expression (15).
TFAM can promote mtDNA transcription using
mitochondrial RNA polymerase in a promoter-specific manner. The
replication of mammalian mtDNA is hypothesized to be coupled with
transcription, and TFAM has been suggested to be essential for the
replication of mtDNA (10–12). TFAM exhibited high expression
levels in colon cancer and upregulated the proliferation of SW480
cells in a previous study (31).
Although the mechanism of TFAM overexpression remains unclear, one
possible mechanism maybe via the regulation of miRNAs. Therefore,
TFAM was selected as the target gene in order to identify the
regulatory miRNA. MiRWalk 2.0 is a comprehensive database providing
information on miRNAs from humans, mice and rats, detailing their
predicted and validated binding sites on their target genes.
MiRWalk summarized the predition frequencies in Microt4, miRanda,
miRBridge, miRDB, miRMap, miRNAMap, PicTar2, PITA, RNA22, RNAhybrid
and TargetScan and the output was a score. Subsequently, miR-590-3p
was selected for further study due to its high prediction score. As
predicted, the present study indicated that TFAM was the direct
target of miR-590-3p in luciferase reporter assays.
In addition, high expression of miRNA-150-3p was
identified in colon cancer tissue, which was positively correlated
with TFAM. It was indicated that miRNA-150-3p may enhance the
expression of TFAM. The overexpression of miRNA-150-3p may promote
the aberrant expression of TFAM, thus contributing to the
progression of colon cancer. In addition, the suppressive effect
against proliferation was confirmed by MTT assays, which
investigated the downregulation of miR-590-3p.
In summary, the present study identified the
interaction of miR-590-3p and TFAM in colon cancer. TFAM was
identified as a target of miR-590-3p, and miR-590-3p enhanced the
proliferation of SW480 cells. miR-590-3p was suggested as a novel
target for knockdown.
References
|
1
|
http://www.who.int/mediacentre/factsheets/fs297/en/Accessed
on 29 August 2012.
|
|
2
|
Bray F, Ren JS, Masuyer E and Ferlay J:
Global estimates of cancer prevalence for 27 sites in the adult
population in 2008. Int J Cancer. 132:1133–1145. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bonawitz ND, Clayton DA and Shadel GS:
Initiation and beyond: Multiple functions of the human
mitochondrial transcription machinery. Mol Cell. 24:813–825. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Parisi MA and Clayton DA: Similarity of
human mitochondrial transcription factor 1 to high mobility group
proteins. Science. 252:965–969. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Litonin D, Sologub M, Shi Y, Savkina M,
Anikin M, Falkenberg M, Gustafsson CM and Temiakov D: Human
mitochondrial transcription revisited: Only TFAM and TFB2M are
required for transcription of the mitochondrial genes in vitro. J
Biol Chem. 285:18129–18133. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Tang D, Kang R, Zeh HJ III and Lotze MT:
High-mobility group box 1 and cancer. Biochim Biophys Acta.
1799:131–140. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Baffy G: Uncoupling protein-2 and cancer.
Mitochondrion. 10:243–252. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yoshida Y, Hasegawa J, Nezu R, Kim YK,
Hirota M, Kawano K, Izumi H and Kohno K: Clinical usefulness of
mitochondrial transcription factor a expression as a predictive
marker in colorectal cancer patients treated with FOLFOX. Cancer
Sci. 102:578–582. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Toki N, Kagami S, Kurita T, Kawagoe T,
Matsuura Y, Hachisuga T, Matsuyama A, Hashimoto H, Izumi H and
Kohno K: Expression of mitochondrial transcription factor A in
endometrial carcinomas: Clinicopathologic correlations and
prognostic significance. Virchows Arch. 456:387–393. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Alam TI, Kanki T, Muta T, Ukaji K, Abe Y,
Nakayama H, Takio K, Hamasaki N and Kang D: Human mitochondrial DNA
is packaged with TFAM. Nucleic Acids Res. 31:1640–1645. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kaufman BA, Durisic N, Mativetsky JM,
Costantino S, Hancock MA, Grutter P and Shoubridge E: The
mitochondrial transcription factor TFAM coordinates the assembly of
multiple DNA molecules into nucleoid-like structures. Mol Biol
Cell. 18:3225–3236. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Xu S, Zhong M, Zhang L, Wang Y, Zhou Z,
Hao Y, Zhang W, Yang X, Wei A, Pei L and Yu Z: Overexpression of
Tfam protects mitochondria against beta-amyloid-induced oxidative
damage in SH-SY5Y cells. FEBS J. 276:3800–3809. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Aydin J, Andersson DC, Hänninen SL,
Wredenberg A, Tavi P, Park CB, Larsson NG, Bruton JD and Westerblad
H: Increased mitochondrial Ca2+ and decreased sarcoplasmic
reticulum Ca2+ in mitochondrial myopathy. Hum Mol Genet.
18:278–288. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kidani A, Izumi H, Yoshida Y, Kashiwagi E,
Ohmori H, Tanaka T, Kuwano M and Kohno K: Thioredoxin2 enhances the
damaged DNA binding activity of mtTFA through direct interaction.
Int J Oncol. 35:1435–1440. 2009.PubMed/NCBI
|
|
15
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Petersen CP, Bordeleau ME, Pelletier J and
Sharp PA: Short RNAs repress translation after initiation in
mammalian cells. Mol Cell. 21:533–542. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Adam L, Zhong M, Choi W, Qi W, Nicoloso M,
Arora A, Calin G, Wang H, Siefker-Radtke A, McConkey D, et al:
MiR-200 expression regulates epithelial-to-mesenchymal transition
in bladder cancer cells and reverses resistance to epidermal growth
factor receptor therapy. Clin Cancer Res. 15:5060–5072. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Saito Y, Suzuki H, Tsugawa H, Nakagawa I,
Matsuzaki J, Kanai Y and Hibi T: Chromatin remodeling at Alu
repeats by epigenetic treatment activates silenced microRNA-512-5p
with down regulation of Mcl-1 in human gastric cancer cells.
Oncogene. 28:2738–2744. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Mazar J, DeYoung K, Khaitan D, Meister E,
Almodovar A, Goydos J, Ray A and Perera RJ: The regulation of
miRNA-211 expression and its role in melanoma cell invasiveness.
PLoS One. 5:e137792010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Di Leva G, Piovan C, Gasparini P, Ngankeu
A, Taccioli C, Briskin D, Cheung DG, Bolon B, Anderlucci L, Alder
H, et al: Estrogen mediated-activation of miR-191/425 cluster
modulates tumorigenicity of breast cancer cells depending on
estrogen receptor status. PLoS Genet. 9:e10033112013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Rivas MA, Venturutti L, Huang YW,
Schillaci R, Huang TH and Elizalde PV: Downregulation of the
tumor-suppressor miR-16 via progestin-mediated oncogenic signaling
contributes to breast cancer development. Breast Cancer Res.
14:R772012. View
Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kalinowski FC, Giles KM, Candy PA, Ali A,
Ganda C, Epis MR, Webster RJ and Leedman PJ: Regulation of
epidermal growth factor receptor signaling and erlotinib
sensitivity in head and neck cancer cells by miR-7. PLoS One.
7:e470672012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Villa C, Fenoglio C, De Riz M, Clerici F,
Marcone A, Benussi L, Ghidoni R, Gallone S, Cortini F, Serpente M,
et al: Role of hnRNP-A1 and miR-590-3p in neuronal death: Genetics
and expression analysis in patients with Alzheimer disease and
frontotemporal lobar degeneration. Rejuvenation Res. 14:275–281.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Vinuesa CG, Rigby RJ and Yu D: Logic and
extent of miRNA-mediated control of autoimmune gene expression. Int
Rev Immunol. 28:112–138. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Benson AB III, Venook AP, Bekaii-Saab T,
Chan E, Chen YJ, Cooper HS, Engstrom PF, Enzinger PC, Fenton MJ,
Fuchs CS, et al: National Comprehensive Cancer Network: Colon
cancer, version 3.2014. J Natl Compr Canc Netw. 12:1028–1059.
2014.PubMed/NCBI
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Belyavsky A, Vinogradova T and Rajewsky K:
PCR-based cDNA library construction: General cDNA libraries at the
level of a few cells. Nucleic Acids Res. 17:2919–2932. 1989.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Esquela-Kerscher A and Slack FJ:
Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer.
6:259–269. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
29
|
Bueno MJ and Malumbres M: MicroRNAs and
the cell cycle. Biochim Biophys Acta. 1812:592–601. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lieberman J, Slack F, Pandolfi PP,
Chinnaiyan A, Agami R and Mendell JT: Noncoding RNAs and cancer.
Cell. 153:9–10. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kai-ming Wu, Yu-long He, Fa-keng Liu and
Jian-hui Chen: Expression of mitochondrial transcription factor a
in colon cancer and its role for proliferative regulation. Chinese
Journal Of Bases And Clinics In General Surgery. 22:576–580.
2015.
|