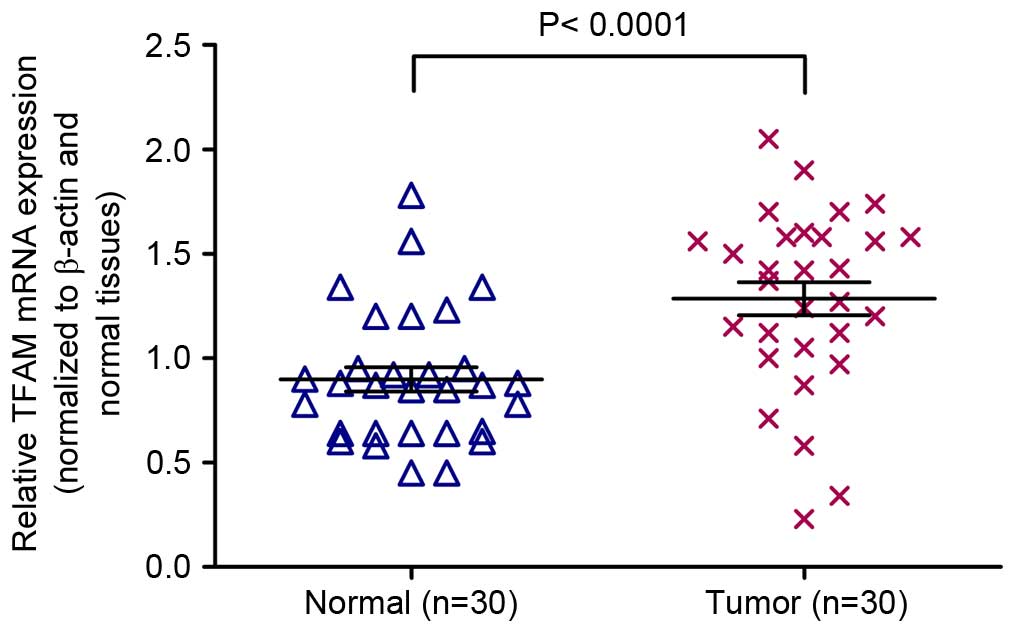
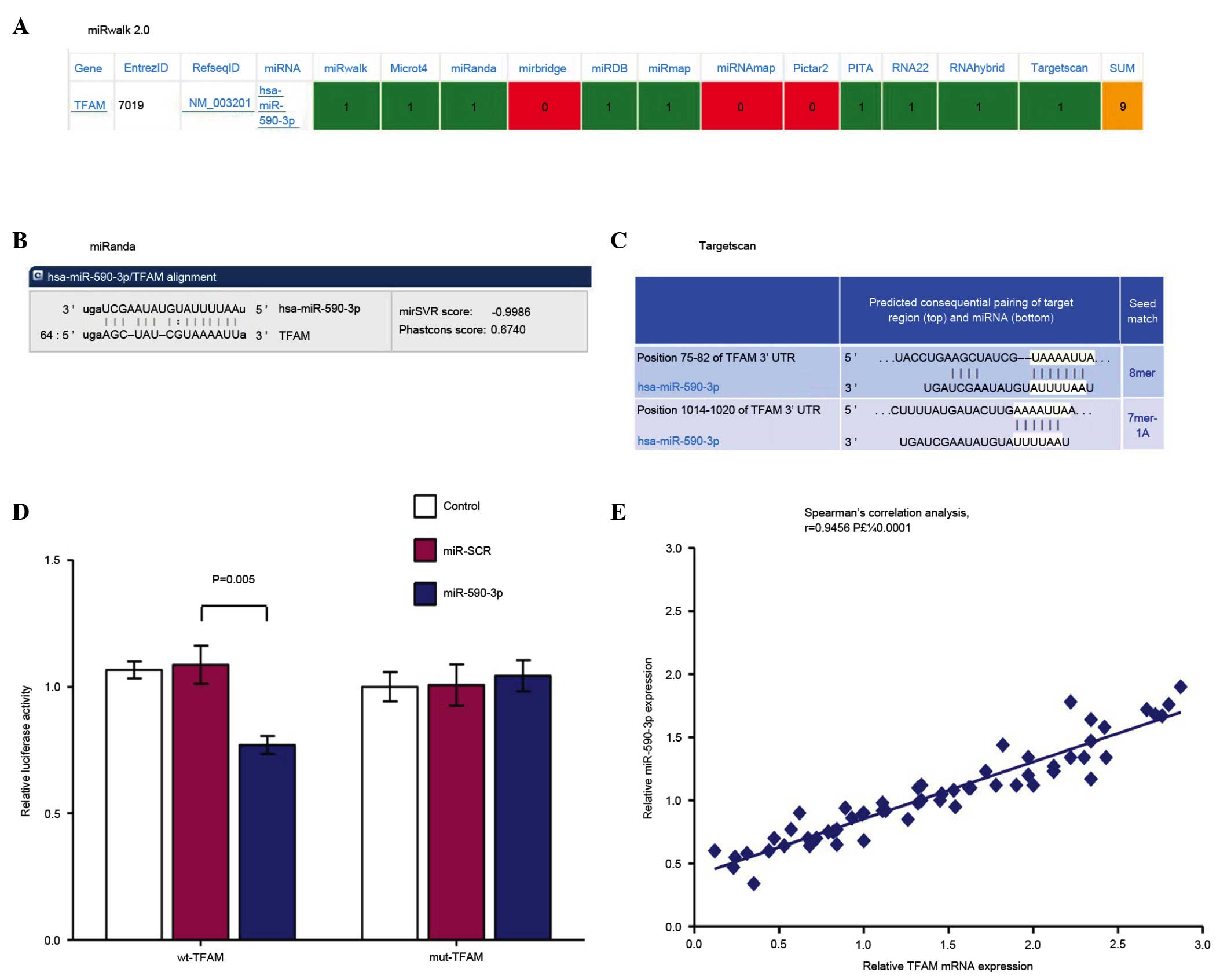
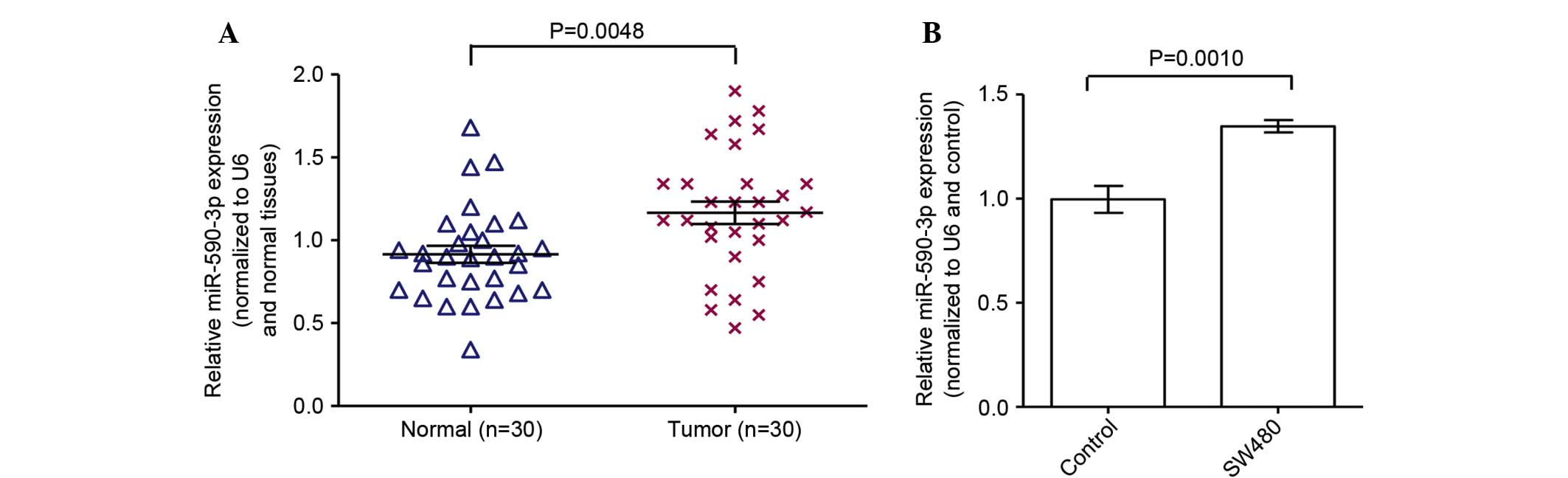
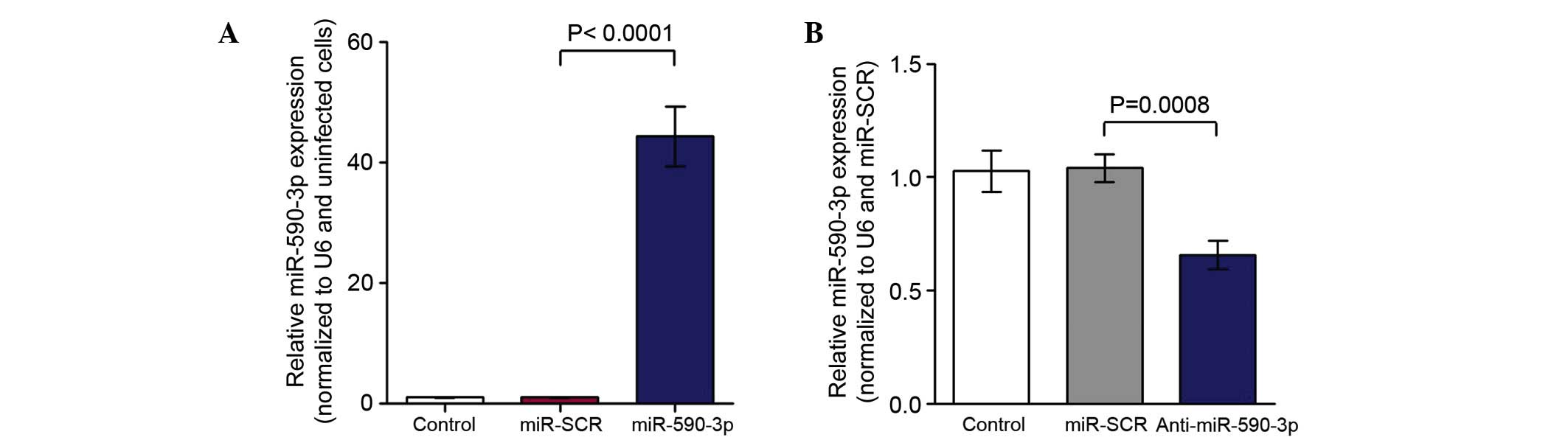
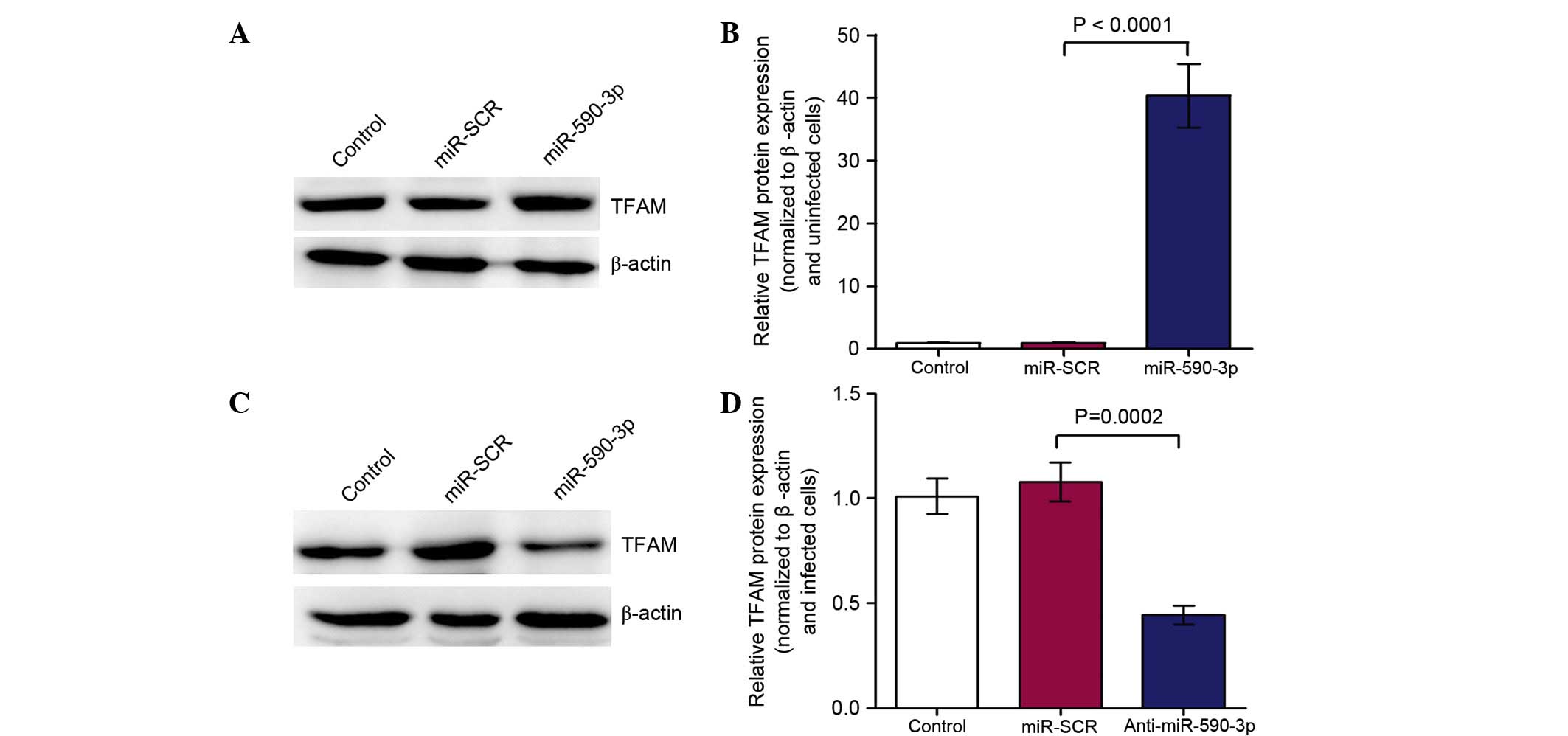
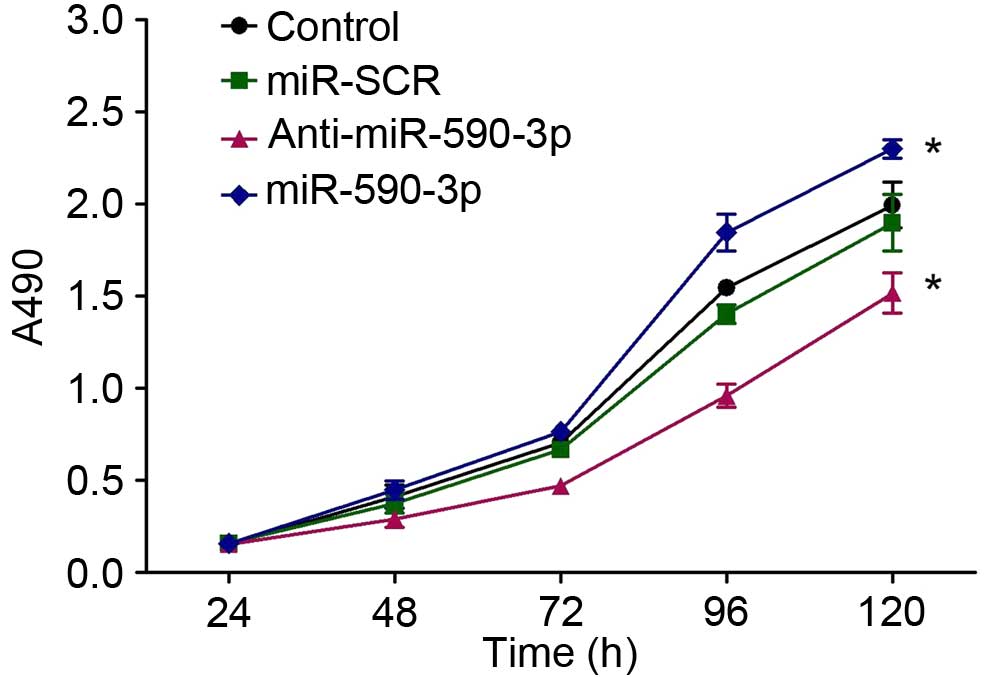
|
1
|
http://www.who.int/mediacentre/factsheets/fs297/en/Accessed
on 29 August 2012.
|
|
2
|
Bray F, Ren JS, Masuyer E and Ferlay J:
Global estimates of cancer prevalence for 27 sites in the adult
population in 2008. Int J Cancer. 132:1133–1145. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bonawitz ND, Clayton DA and Shadel GS:
Initiation and beyond: Multiple functions of the human
mitochondrial transcription machinery. Mol Cell. 24:813–825. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Parisi MA and Clayton DA: Similarity of
human mitochondrial transcription factor 1 to high mobility group
proteins. Science. 252:965–969. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Litonin D, Sologub M, Shi Y, Savkina M,
Anikin M, Falkenberg M, Gustafsson CM and Temiakov D: Human
mitochondrial transcription revisited: Only TFAM and TFB2M are
required for transcription of the mitochondrial genes in vitro. J
Biol Chem. 285:18129–18133. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Tang D, Kang R, Zeh HJ III and Lotze MT:
High-mobility group box 1 and cancer. Biochim Biophys Acta.
1799:131–140. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Baffy G: Uncoupling protein-2 and cancer.
Mitochondrion. 10:243–252. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yoshida Y, Hasegawa J, Nezu R, Kim YK,
Hirota M, Kawano K, Izumi H and Kohno K: Clinical usefulness of
mitochondrial transcription factor a expression as a predictive
marker in colorectal cancer patients treated with FOLFOX. Cancer
Sci. 102:578–582. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Toki N, Kagami S, Kurita T, Kawagoe T,
Matsuura Y, Hachisuga T, Matsuyama A, Hashimoto H, Izumi H and
Kohno K: Expression of mitochondrial transcription factor A in
endometrial carcinomas: Clinicopathologic correlations and
prognostic significance. Virchows Arch. 456:387–393. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Alam TI, Kanki T, Muta T, Ukaji K, Abe Y,
Nakayama H, Takio K, Hamasaki N and Kang D: Human mitochondrial DNA
is packaged with TFAM. Nucleic Acids Res. 31:1640–1645. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kaufman BA, Durisic N, Mativetsky JM,
Costantino S, Hancock MA, Grutter P and Shoubridge E: The
mitochondrial transcription factor TFAM coordinates the assembly of
multiple DNA molecules into nucleoid-like structures. Mol Biol
Cell. 18:3225–3236. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Xu S, Zhong M, Zhang L, Wang Y, Zhou Z,
Hao Y, Zhang W, Yang X, Wei A, Pei L and Yu Z: Overexpression of
Tfam protects mitochondria against beta-amyloid-induced oxidative
damage in SH-SY5Y cells. FEBS J. 276:3800–3809. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Aydin J, Andersson DC, Hänninen SL,
Wredenberg A, Tavi P, Park CB, Larsson NG, Bruton JD and Westerblad
H: Increased mitochondrial Ca2+ and decreased sarcoplasmic
reticulum Ca2+ in mitochondrial myopathy. Hum Mol Genet.
18:278–288. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kidani A, Izumi H, Yoshida Y, Kashiwagi E,
Ohmori H, Tanaka T, Kuwano M and Kohno K: Thioredoxin2 enhances the
damaged DNA binding activity of mtTFA through direct interaction.
Int J Oncol. 35:1435–1440. 2009.PubMed/NCBI
|
|
15
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Petersen CP, Bordeleau ME, Pelletier J and
Sharp PA: Short RNAs repress translation after initiation in
mammalian cells. Mol Cell. 21:533–542. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Adam L, Zhong M, Choi W, Qi W, Nicoloso M,
Arora A, Calin G, Wang H, Siefker-Radtke A, McConkey D, et al:
MiR-200 expression regulates epithelial-to-mesenchymal transition
in bladder cancer cells and reverses resistance to epidermal growth
factor receptor therapy. Clin Cancer Res. 15:5060–5072. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Saito Y, Suzuki H, Tsugawa H, Nakagawa I,
Matsuzaki J, Kanai Y and Hibi T: Chromatin remodeling at Alu
repeats by epigenetic treatment activates silenced microRNA-512-5p
with down regulation of Mcl-1 in human gastric cancer cells.
Oncogene. 28:2738–2744. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Mazar J, DeYoung K, Khaitan D, Meister E,
Almodovar A, Goydos J, Ray A and Perera RJ: The regulation of
miRNA-211 expression and its role in melanoma cell invasiveness.
PLoS One. 5:e137792010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Di Leva G, Piovan C, Gasparini P, Ngankeu
A, Taccioli C, Briskin D, Cheung DG, Bolon B, Anderlucci L, Alder
H, et al: Estrogen mediated-activation of miR-191/425 cluster
modulates tumorigenicity of breast cancer cells depending on
estrogen receptor status. PLoS Genet. 9:e10033112013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Rivas MA, Venturutti L, Huang YW,
Schillaci R, Huang TH and Elizalde PV: Downregulation of the
tumor-suppressor miR-16 via progestin-mediated oncogenic signaling
contributes to breast cancer development. Breast Cancer Res.
14:R772012. View
Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kalinowski FC, Giles KM, Candy PA, Ali A,
Ganda C, Epis MR, Webster RJ and Leedman PJ: Regulation of
epidermal growth factor receptor signaling and erlotinib
sensitivity in head and neck cancer cells by miR-7. PLoS One.
7:e470672012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Villa C, Fenoglio C, De Riz M, Clerici F,
Marcone A, Benussi L, Ghidoni R, Gallone S, Cortini F, Serpente M,
et al: Role of hnRNP-A1 and miR-590-3p in neuronal death: Genetics
and expression analysis in patients with Alzheimer disease and
frontotemporal lobar degeneration. Rejuvenation Res. 14:275–281.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Vinuesa CG, Rigby RJ and Yu D: Logic and
extent of miRNA-mediated control of autoimmune gene expression. Int
Rev Immunol. 28:112–138. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Benson AB III, Venook AP, Bekaii-Saab T,
Chan E, Chen YJ, Cooper HS, Engstrom PF, Enzinger PC, Fenton MJ,
Fuchs CS, et al: National Comprehensive Cancer Network: Colon
cancer, version 3.2014. J Natl Compr Canc Netw. 12:1028–1059.
2014.PubMed/NCBI
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Belyavsky A, Vinogradova T and Rajewsky K:
PCR-based cDNA library construction: General cDNA libraries at the
level of a few cells. Nucleic Acids Res. 17:2919–2932. 1989.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Esquela-Kerscher A and Slack FJ:
Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer.
6:259–269. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
29
|
Bueno MJ and Malumbres M: MicroRNAs and
the cell cycle. Biochim Biophys Acta. 1812:592–601. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lieberman J, Slack F, Pandolfi PP,
Chinnaiyan A, Agami R and Mendell JT: Noncoding RNAs and cancer.
Cell. 153:9–10. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kai-ming Wu, Yu-long He, Fa-keng Liu and
Jian-hui Chen: Expression of mitochondrial transcription factor a
in colon cancer and its role for proliferative regulation. Chinese
Journal Of Bases And Clinics In General Surgery. 22:576–580.
2015.
|