Introduction
MicroRNAs (miRNAs) are a class of small non-coding
RNAs that have been extensively studied as crucial negative
regulatory molecules. Canonical miRNA is first processed from a
primary miRNA (pri-miRNA) transcript via the Drosha-dependent
microprocessor complex (1), and
the process generates a precursor miRNA (pre-miRNA) with a
stem-loop hairpin structure. Then, it is recognized and cut by
Dicer into miRNA-miRNA* duplexes (2) and subsequently pre-miRNA is
transported to the cytoplasm by exportin-5 (3). The Dicer protein is widely
distributed in plants, metazoans and fungi (4,5), and
it is also involved in the processing of other small RNA species,
including small nuclear RNAs (6).
Recurrent somatic mutations in Drosha can induce changes in miRNA
expression (7), and somatic
mutations in Drosha and Dicer1 can impair miRNA biogenesis
(8). Simultaneously, alternative
pathways of miRNA biogenesis have been reported (9), and some miRNAs are generated by a
novel processing pathway independent of Dicer (10–13).
For example, mirtrons can be processed by splicing from precursor
hairpins (14,15), and their precursor sequences are
shorter than canonical pri-miRNAs because these comprise
miRNA-miRNA* duplexes alone. Pre-miRNA hairpins of miRNAs from
mirtron-like sources are generated via splicing of short introns
and exosome-mediated trimming (16,17).
Typical miRNA biogenesis indicates that mature
miRNAs can be incorporated into the RNA-induced silencing complex
that then binds to the 3′-untranslated region of the target mRNA to
degrade mRNAs or to repress translation (18), while another strand, termed miRNA
star (miRNA*), is degraded to an inactive strand. However,
accumulating evidence suggests that degraded miRNA* may also serve
an important role in gene regulation at the post-transcriptional
levels, as well as in the mature miRNA sequence (19–23),
and miRNA-miRNA* duplexes are also termed miRNA (miR)-#-5p-miR-#-3p
duplexes. In the miRNA locus, a series of miRNA variants, called
isomiRs, have been widely detected based on high-throughput
sequencing datasets (24–30). These multiple isomiRs are
predominantly derived from the alternative and imprecise cleavage
of Drosha and Dicer during pri-miRNA/pre-miRNA processing, and 3′
addition events in miRNA maturation processes (27). In the specific miRNA locus, isomiR
profiles are always stable across different samples and different
species (26,30), indicating a relatively stable miRNA
maturation process.
Drosha and Dicer have important roles in the miRNA
maturation process, and their imprecise and alternative cleavage
processes largely contribute to the generation of multiple isomiRs.
It is known that the isomiR profiles are always stable because of
Drosha and Dicer, even across different animal species, however
fewer studies have focused on isomiR expression profiles between
typical miRNA loci and Dicer-independent miRNA loci. Generally,
several dominant isomiRs (typically one to three) can be identified
in the canonical miRNA locus, and others always possess lower
expression rates (26,29,31).
Therefore, according to characteristics of cleavage and expression
patterns of multiple isomiRs, the current study attempted to
explore the potential associations between isomiR profiles in
different miRNA loci with diversity in the maturation processes,
and simultaneously discuss evolutionary patterns of Drosha/Dicer.
According to miRNA expression profiles, abundantly expressed
miR-451 has been identified as an miRNA that is Dicer-independent,
and based on previous studies focusing on isomiR expression
(32–34), relevant expression and evolutionary
analysis were performed using public datasets. The present study
may provide additional information regarding miRNA/isomiR
biogenesis.
Materials and methods
Source data
Both Drosha and Dicer nucleotide and protein
sequences of eight vertebrates, including Amphibia (Xenopus
tropicalis), Aves (Gallus gallus; Taeniopygia guttata), Mammalia
(Homo sapiens; Sus scrofa; Mus musculus), Pisces (Danio rerio) and
Sauria (Anolis carolinensis) were collected from GenBank (Table I). Simultaneously, available
expression data of miRNA/isomiR in breast cancer (BC) samples
(n=683) and normal samples (n=87) were acquired from the Cancer
Genome Atlas pilot project (http://cancergenome.nih.gov). Then, due to the fact
that the selected BC samples were derived from female patients,
samples from other human diseases from the Cancer Genome Atlas
pilot project (http://cancergenome.nih.gov) were selected: Thyroid
carcinoma (tumor, n=507; normal, n=59) and prostate adenocarcinoma
(tumor, n=498; normal, n=52). The selection generated more
information about isomiR expression with potential gender
differences.
 | Table I.Drosha and Dicer in eight
vertebrates. |
Table I.
Drosha and Dicer in eight
vertebrates.
| Species | Abbreviation | Gene | Accession number | Length (AA) |
|---|
| Amphibia: Xenopus
tropicalis | xtr | Drosha | NP_001107152.1 | 1,325 |
| Aves: Gallus
gallus | gga | Drosha | NP_001006379.1 | 1,336 |
| Aves:
Taeniopygia guttata | tgu | Drosha | XP_002199233 | 1,338 |
| Mammalia: Homo
sapiens | hsa | Drosha | Q9NRR4 | 1,374 |
| Mammalia: Sus
scrofa | ssc | Drosha | XP_005672458 | 1,373 |
| Mammalia: Mus
musculus |
mmu | Drosha | Q5HZJ0 | 1,373 |
| Pisces: Danio
rerio | dre | Drosha | NP_001103942 | 1,289 |
| Sauria: Anolis
carolinensis | aca | Drosha | XP_003226799 | 1,339 |
| Amphibia:
Xenopus tropicalis | xtr | Dicer | NP_001123390.2 | 1,893 |
| Aves: Gallus
gallus | gga | Dicer | NP_001035555.1 | 1,921 |
| Aves:
Taeniopygia guttata | tgu | Dicer | NP_001156875 | 1,921 |
| Mammalia: Homo
sapiens | hsa | Dicer | NP_001278557.1 | 1,922a |
| Mammalia: Sus
scrofa | ssc | Dicer | NP_001184123.1 | 1,915 |
| Mammalia: Mus
musculus |
mmu | Dicer | NP_683750.2 | 1,906 |
| Pisces: Danio
rerio | dre | Dicer | NP_001154925.1 | 1,865 |
| Sauria: Anolis
carolinensis | aca | Dicer | XP_003214365 | 1,918 |
Sequence and expression analyses
The amino acid sequences of Drosha and Dicer were
aligned using Clustal X 2.0 (http://www.clustal.org/clustal2) (35), and phylogenetic associations were
reconstructed using the MEGA 5.10 software (http://www.megasoftware.net) (36) based on the neighbor-joining method.
Genetic distance was simultaneously estimated using MEGA.
Functional domains of Drosha and Dicer were identified using the
Pfam database (http://pfam.xfam.org) (37), and inferred domains were predicted
using SMART (http://smart.embl-heidelberg.de) (38).
According to biological characteristics of isomiRs,
the relative expression rate of each isomiR was estimated based on
all the isomiRs in the miRNA locus, and then the average expression
rate was calculated across different individuals. Herein, due to
the larger differences in sample sizes between tumor and normal
samples, expression patterns were first estimated using different
sample sizes, and then were estimated using equal sample sizes by
randomly selecting 87 tumor samples in BC. Simultaneously, relevant
analysis was further performed using other samples. According to
miRNA/isomiR expression profiles and typical isomiR expression
patterns, the simultaneously dominantly expressed miR-#-5p and
miR-#-3p from hsa-miR-21, −30a and −30e were collected, and another
miRNA that was Dicer-independent (miR-451) was selected for further
analysis.
Statistical analysis
The expression levels of the isomiRs in multiple
individuals, particularly those of dominant isomiRs, were expressed
as the mean ± standard deviation. Differences in genetic distance
were estimated using the t-test between Drosha and Dicer, and
P<0.05 was considered to indicate a statistically significant
difference. Simultaneously, the 95% confidence interval was also
estimated. The degree of variation of relative expression of
isomiRs was presented using a box plot. Relevant statistical
description or analysis was conducted using Stata software, version
11 (StataCorp LP, College Station, TX, USA).
Results and discussion
The typical miRNA maturation process depends on
Drosha and Dicer, and the miRNA-#-5p:miRNA-#-3p duplex is generated
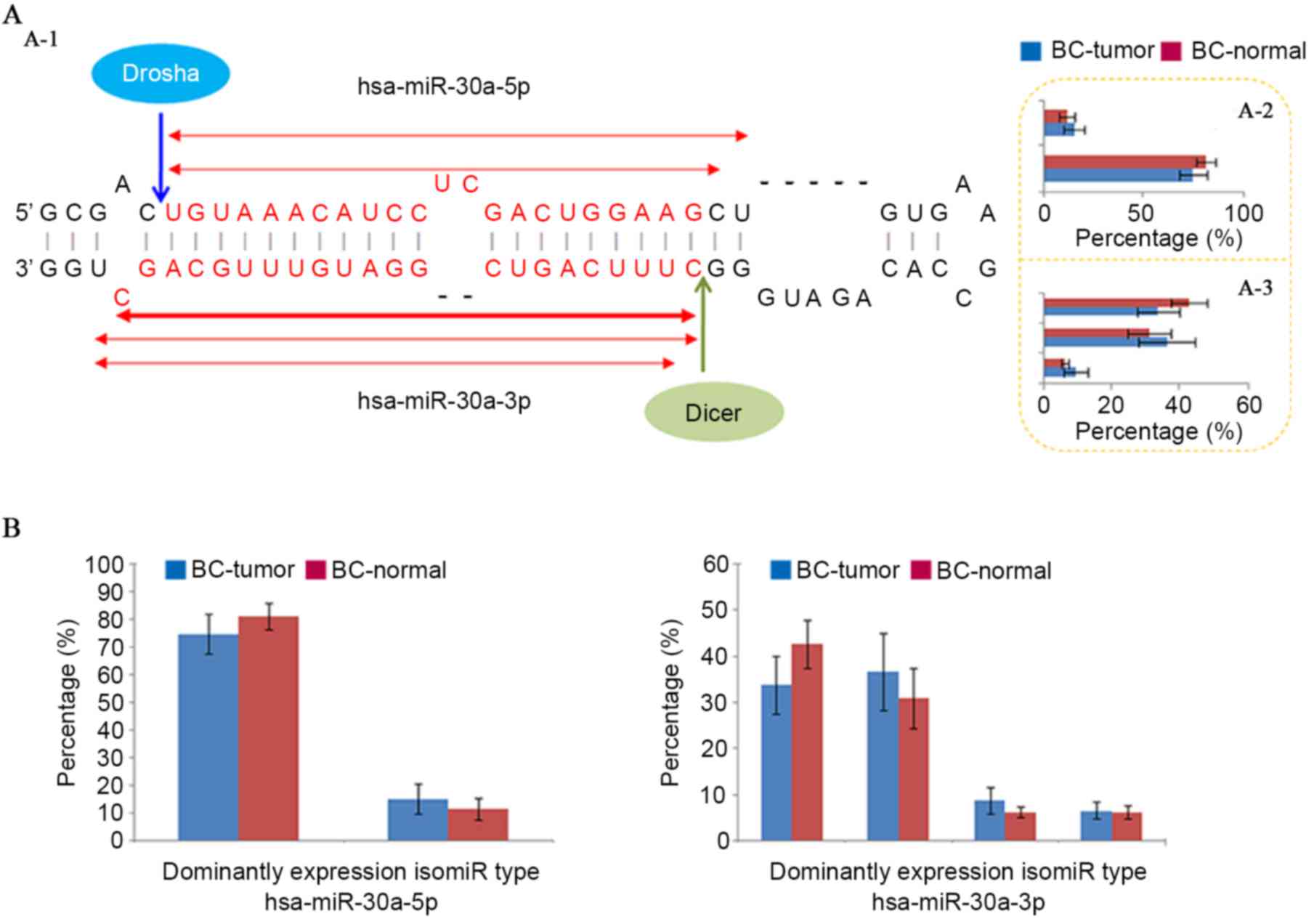
through cleavage of pri-miRNA and pre-miRNA (Fig. 1A). Although single miRNA sequences
have been widely studied, the miRNA locus generates a series of
isomiRs with diverse sequences and expression levels that are
predominantly derived from the imprecise and alternative cleavage
of Drosha/Dicer and 3′ addition events (24–30).
As a specific sequence in multiple isomiRs, the canonical miRNA
sequence is not always the most dominantly expressed sequence (such
as hsa-miR-30a-5p in the miRNA locus; Fig. 1) (29). Although sample sizes differed
between tumor and normal samples, similar expression patterns were
obtained using equally sized samples (Fig. 1). The isomiR expression profiles
were always stable across different samples, and this feature was
also observed in different cells, tissues and animal species,
although abnormal isomiR expression patterns were also detected,
particularly in certain diseased samples. The current study
attempted to compare isomiR expression profiles between typical
miRNA loci and miRNA loci with independent of Dicer using public
sequencing datasets. Compared with recent studies on the expression
and evolutionary patterns of isomiRs (29,39–41),
the present study examined the expression profile of a specific
miRNA gene (hsa-miR-30a), that can generate two kinds of mature
miRNAs, and a dominantly expressed miRNA independent of Dicer
(hsa-miR-451), which has been updated as miR-451a, and was selected
due to the fact that fewer miRNAs independent of Dicer are
dominantly expressed.
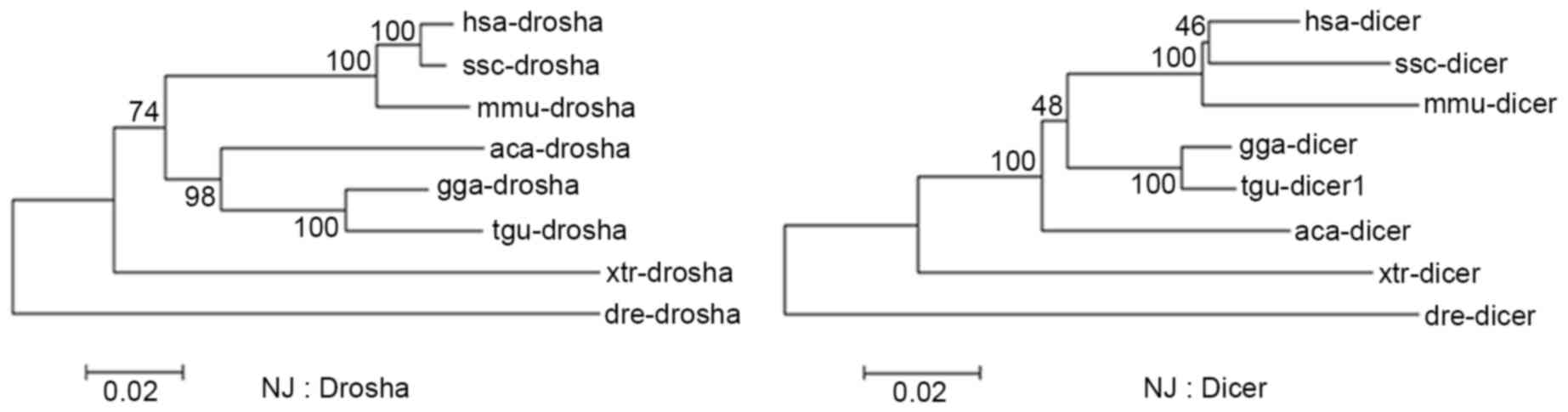
As crucial factors that lead to multiple isomiRs in
miRNA maturation, Drosha and Dicer are well-conserved and have
similar evolutionary rates across different vertebrates (Fig. 2 and Table II). These similar evolutionary
patterns may contribute to recognition and cleavage of Drosha and
Dicer in the miRNA maturation process, and further contribute to
stable isomiR expression profiles, particularly for the majority of
isomiRs that are 3′ isomiRs with the same 5′ ends. In pre-miRNA
with a stem-loop structure, dominantly expressed miRNA may be
located in the 5p or 3p arm, however all the miRNA loci generate
dominant 3′ isomiRs and rare 5′ isomiRs. Notably, more pre-miRNAs
have been identified to generate mature and functional miR-#-5p and
miR-#-3p. These observations suggest the relative precise cleavage
of Drosha and Dicer on the 5′ ends during miRNA maturation process,
although they serve important roles in different regions in cells.
Divergence of 3′ ends among isomiRs from a given miRNA locus and
among miRNAs in different species may be predominantly derived from
the imprecise cleavage of Drosha and Dicer on 3′ ends, in addition
to modification events following miRNA maturation.
 | Table II.Evolutionary distance between Drosha
and Dicer. |
Table II.
Evolutionary distance between Drosha
and Dicer.
|
| hsa | ssc | mmu | gga | tgu | aca | xtr | dre |
|---|
| hsa | – | 0.048 | 0.055 | 0.066 | 0.068 | 0.091 | 0.150 | 0.200 |
| ssc | 0.012 | – | 0.071 | 0.089 | 0.088 | 0.106 | 0.162 | 0.201 |
| mmu | 0.033 | 0.036 | – | 0.089 | 0.090 | 0.112 | 0.165 | 0.220 |
| gga | 0.112 | 0.111 | 0.115 | – | 0.019 | 0.071 | 0.134 | 0.199 |
| tgu | 0.121 | 0.119 | 0.127 | 0.045 | – | 0.070 | 0.138 | 0.200 |
| aca | 0.126 | 0.122 | 0.132 | 0.095 | 0.109 | – | 0.144 | 0.198 |
| xtr | 0.180 | 0.178 | 0.178 | 0.160 | 0.170 | 0.167 | – | 0.213 |
| dre | 0.204 | 0.203 | 0.208 | 0.212 | 0.226 | 0.226 | 0.242 | – |
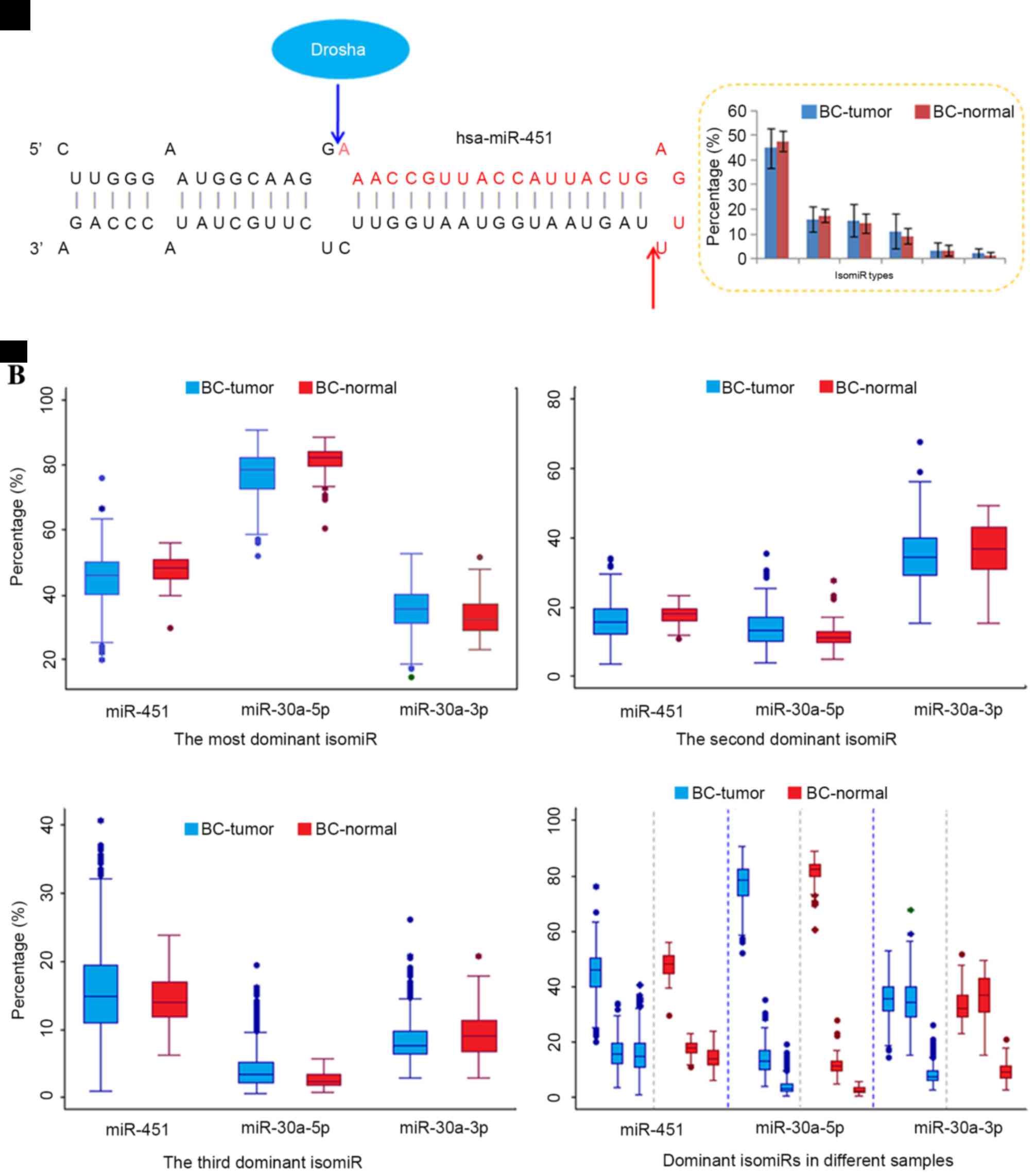
However, compared with typical miRNAs that are
generated by cleavage of Drosha and Dicer, certain miRNAs are
generated by alternative pathways during miRNA biogenesis (9), including a novel processing pathway
independent of Dicer (10–12). For example, miR-451 is generated
via the Dicer-independent pathway, and then contributes to the
regulatory network as a functional miRNA (13). Although different maturation
processes occur in miR-451 and canonical miRNA loci (including
miR-21, 30a and 30e), multiple isomiR sequences are also present in
the Dicer-independent miRNA locus (Figs. 3–5
and Table III). Similar to
typical miRNA loci (26,29,42),
several dominant isomiRs are dominantly expressed, and the majority
of isomiRs are 3′ isomiRs. The highly conserved 5′ ends ensure the
consistency of the function between different isomiR sequences,
which in turn facilitate the co-regulation of target mRNAs.
However, the most dominant isomiR only possesses moderate
expression (approximately 45% of the total expression), and other
dominant isomiRs have similar expression levels (Fig. 3A). The moderate expression patterns
differ from the most typical miRNAs that are prone to have
predominant isomiRs with absolute abundant expression (such as
isomiRs from the miR-30a-5p locus).
 | Table III.Dominantly expressed isomiR sequences
in Figs. 2 and 3. |
Table III.
Dominantly expressed isomiR sequences
in Figs. 2 and 3.
| miRNA | IsomiR species | Sequence |
|---|
| miR-451 | The most
dominant |
AAACCGUUACCAUUACUGAGUU |
|
| The second
dominant |
AAACCGUUACCAUUACUGAGU |
|
| The third
dominant |
AAACCGUUACCAUUACUGAGUUU |
| miR-30a-5p | The most
dominant |
UGUAAACAUCCUCGACUGGAAGC |
|
| The second
dominant |
UGUAAACAUCCUCGACUGGAAGCU |
|
| The third
dominant |
UGUAAACAUCCUCGACUGGAAG |
| miR-30a-3p | The most
dominant |
CUUUCAGUCGGAUGUUUGCAGCU |
|
| The second
dominant |
CUUUCAGUCGGAUGUUUGCAGC |
|
| The third
dominant |
UUUCAGUCGGAUGUUUGCAGCU |
| miR-21-5p | The most
dominant |
UAGCUUAUCAGACUGAUGUUGAC |
|
| The second
dominant |
UAGCUUAUCAGACUGAUGUUGA |
|
| The third
dominant |
UAGCUUAUCAGACUGAUGUUGACU |
| miR-21-3p | The most
dominant |
CAACACCAGUCGAUGGGCUGUCU |
|
| The second
dominant |
CAACACCAGUCGAUGGGCUGUC |
|
| The third
dominant |
CAACACCAGUCGAUGGGCUGU |
| miR-30e-5p | The most
dominant |
UGUAAACAUCCUUGACUGGA |
|
| The second
dominant |
UGUAAACAUCCUUGACUGGAAGC |
|
| The third
dominant |
UGUAAACAUCCUUGACUGGAAGCU |
| miR-30e-3p | The most
dominant |
CUUUCAGUCGGAUGUUUACAGCG |
|
| The second
dominant |
CUUUCAGUCGGAUGUUUACAGC |
|
| The third
dominant |
CUUUCAGUCGGAUGUUUACAG |
Another non-dominant miRNA strand such as the
miR-30a-3p locus induces moderate isomiR expression profiles,
similar to that observed in Dicer-independent miR-451 locus
(40). Notably, although several
dominant isomiRs are present in the passenger strand, distinct
differences were detected between dominant isomiRs and other rare
isomiRs, whereas in the miR-451 locus, no evident boundary was
detected. The moderate level of expression may have resulted from
the random cleavage of the hairpin, whereas isomiRs in most typical
miRNAs are strictly controlled. The mir-451 hairpin could not be
recognized by Dicer because of the insufficient amount of duplexes
formed, and therefore it was directly cleaved by Ago2. The
Dicer-independent maturation process may lead to relative random
cleavage sites at the 3′ ends of miRNAs, which is different from
the non-random 3′ ends of multiple isomiRs derived from typical
miRNA locus. Generally, in typical miRNA loci, dominant isomiRs are
evident, and there are always one to three types of abundantly
expressed isomiRs that can be used to infer dominant cleavage sites
in Drosha and Dicer. These observations indicate the cleavage bias
of Drosha and Dicer, which further contributes to the distinct
expression profiles of dominant isomiRs.
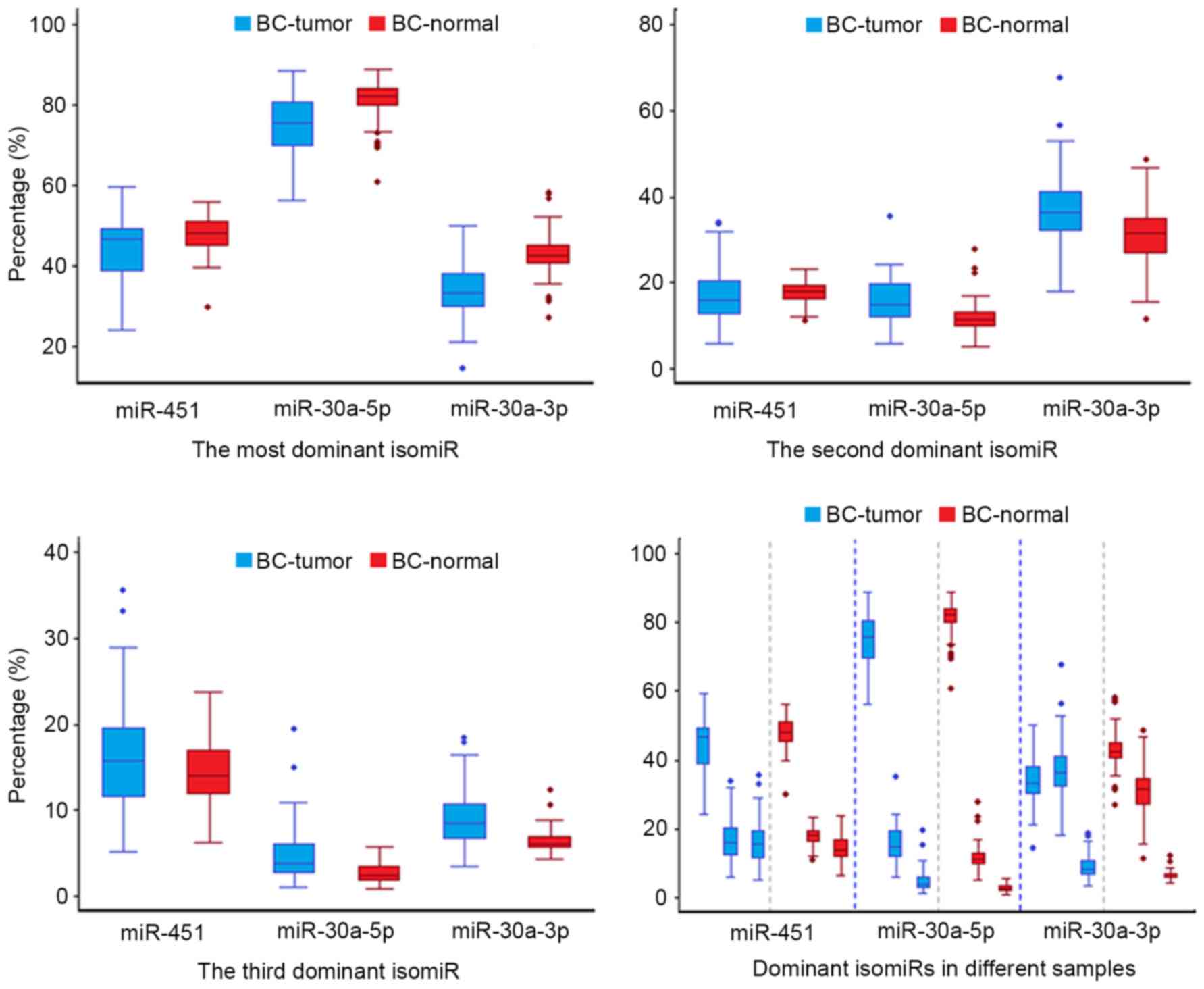
The current study also observed that although isomiR
expression profiles are always stable across different samples,
including between tumor and normal samples, and variations were
also detected between diseased and normal samples (39). In diseased samples, a larger degree
of variation was observed compared with that among normal samples,
which implicates that the miRNA maturation process may be affected
in the abnormal micro-environment (Figs. 3 and 5). The dispersed expression patterns of
isomiRs may be derived from changes in Drosha and Dicer cleavage,
modification or regulation following miRNA maturation in tumor
samples. These distinct characteristics may furthermore provide
additional information on the dynamic expression profiles of the
coding-non-coding RNA regulatory network.
In conclusion, expression analysis of isomiRs
demonstrated that Dicer-independent miRNA involves a moderate level
of isomiR expression compared with that observed in typical miRNA
loci. Notably, a similar expression profile can be detected between
non-dominant miRNA and Dicer-independent miRNA loci, thereby
suggesting more complex miRNA maturation processes, particularly at
the isomiR levels. Dynamic miRNA/isomiR expression profiles further
enrich the regulatory network, particularly those involving
miRNA-miRNA and miRNA-mRNA interactions. Additional studies on
multiple isomiRs, particularly their potential versatile biological
roles, may prove beneficial.
Acknowledgements
The current study was supported by the National
Natural Science Foundation of China (grant nos. 31271261, 31401009
and 61301251), the Program for the Top Young Talents by the
Organization Department of the CPC Central Committee, the National
Natural Science Foundation of Jiangsu (grant no. BK20130885), the
Key Project of Social Development in Jiangsu Province (grant no.
BE2016773), the Research Fund for the Doctoral Program of Higher
Education of China (grant no. 20133234120009), Shandong Provincial
Key Laboratory of Functional Macromolecular Biophysics, the
Priority Academic Program Development of Jiangsu Higher Education
Institutions (PAPD), sponsored by NUPTSF (grant no. NY215068).
References
|
1
|
Gregory RI, Yan KP, Amuthan G, Chendrimada
T, Doratotaj B, Cooch N and Shiekhattar R: The Microprocessor
complex mediates the genesis of microRNAs. Nature. 432:235–240.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
He L and Hannon GJ: MicroRNAs: Small RNAs
with a big role in gene regulation. Nat Rev Genet. 5:522–531. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lund E, Güttinger S, Calado A, Dahlberg JE
and Kutay U: Nuclear export of microRNA precursors. Science.
303:95–98. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Cerutti H and Casas-Mollano JA: On the
origin and functions of RNA-mediated silencing: From protists to
man. Curr Genet. 50:81–99. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Shabalina SA and Koonin EV: Origins and
evolution of eukaryotic RNA interference. Trends Ecol Evol.
23:578–587. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Langenberger D, Bermudez-Santana CI,
Stadler PF and Hoffmann S: Identification and classification of
small RNAs in transcriptome sequence data. Pac Symp Biocomput.
80–87. 2010.PubMed/NCBI
|
|
7
|
Torrezan GT, Ferreira EN, Nakahata AM,
Barros BD, Castro MT, Correa BR, Krepischi AC, Olivieri EH, Cunha
IW, Tabori U, et al: Recurrent somatic mutation in DROSHA induces
microRNA profile changes in Wilms tumour. Nat Commun. 5:40392014.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Rakheja D, Chen KS, Liu Y, Shukla AA,
Schmid V, Chang TC, Khokhar S, Wickiser JE, Karandikar NJ, Malter
JS, et al: Somatic mutations in DROSHA and DICER1 impair microRNA
biogenesis through distinct mechanisms in Wilms tumours. Nat
Commun. 2:48022014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yang JS and Lai EC: Alternative miRNA
biogenesis pathways and the interpretation of core miRNA pathway
mutants. Mol Cell. 43:892–903. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Cheloufi S, Dos Santos CO, Chong MM and
Hannon GJ: A dicer-independent miRNA biogenesis pathway that
requires Ago catalysis. Nature. 465:584–589. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Cifuentes D, Xue H, Taylor DW, Patnode H,
Mishima Y, Cheloufi S, Ma E, Mane S, Hannon GJ, Lawson ND, et al: A
novel miRNA processing pathway independent of Dicer requires
Argonaute2 catalytic activity. Science. 328:1694–1698. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Miyoshi K, Miyoshi T and Siomi H: Many
ways to generate microRNA-like small RNAs: Non-canonical pathways
for microRNA production. Mol Genet Genomics. 284:95–103. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yang JS, Maurin T, Robine N, Rasmussen KD,
Jeffrey KL, Chandwani R, Papapetrou EP, Sadelain M, O'Carroll D and
Lai EC: Conserved vertebrate mir-451 provides a platform for
Dicer-independent, Ago2-mediated microRNA biogenesis. Proc Natl
Acad Sci USA. 107:15163–15168. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Okamura K, Hagen JW, Duan H, Tyler DM and
Lai EC: The mirtron pathway generates microRNA-class regulatory
RNAs in Drosophila. Cell. 130:89–100. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ruby JG, Jan CH and Bartel DP: Intronic
microRNA precursors that bypass Drosha processing. Nature.
448:83–86. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Flynt AS, Greimann JC, Chung WJ, Lima CD
and Lai EC: MicroRNA biogenesis via splicing and exosome-mediated
trimming in Drosophila. Mol Cell. 38:900–907. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chong MM, Zhang G, Cheloufi S, Neubert TA,
Hannon GJ and Littman DR: Canonical and alternate functions of the
microRNA biogenesis machinery. Genes Dev. 24:1951–1960. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kim VN, Han J and Siomi MC: Biogenesis of
small RNAs in animals. Nat Rev Mol Cell Bio. 10:126–139. 2009.
View Article : Google Scholar
|
|
19
|
Okamura K, Phillips MD, Tyler DM, Duan H,
Chou YT and Lai EC: The regulatory activity of microRNA* species
has substantial influence on microRNA and 3′ UTR evolution. Nat
Struct Mol Biol. 15:354–363. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Czech B, Zhou R, Erlich Y, Brennecke J,
Binari R, Villalta C, Gordon A, Perrimon N and Hannon GJ:
Hierarchical rules for Argonaute loading in Drosophila. Mol Cell.
36:445–456. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Okamura K, Liu N and Lai EC: Distinct
mechanisms for microRNA strand selection by Drosophila Argonautes.
Mol Cell. 36:431–444. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Guo L and Lu Z: The fate of miRNA* strand
through evolutionary analysis: Implication for degradation as
merely carrier strand or potential regulatory molecule? PLoS One.
5:e113872010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Jagadeeswaran G, Zheng Y, Sumathipala N,
Jiang H, Arrese EL, Soulages JL, Zhang W and Sunkar R: Deep
sequencing of small RNA libraries reveals dynamic regulation of
conserved and novel microRNAs and microRNA-stars during silkworm
development. BMC Genomics. 11:522010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Landgraf P, Rusu M, Sheridan R, Sewer A,
Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M,
et al: A mammalian microRNA expression atlas based on small RNA
library sequencing. Cell. 129:1401–1414. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Morin RD, Aksay G, Dolgosheina E, Ebhardt
HA, Magrini V, Mardis ER, Sahinalp SC and Unrau PJ: Comparative
analysis of the small RNA transcriptomes of Pinus contorta and
Oryza sativa. Genome Res. 18:571–584. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Guo L, Yang Q, Lu J, Li H, Ge Q, Gu W, Bai
Y and Lu Z: A comprehensive survey of mirna repertoire and 3′
addition events in the placentas of patients with pre-eclampsia
from high-throughput sequencing. PLoS One. 6:e210722011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Neilsen CT, Goodall GJ and Bracken CP:
IsomiRs-the overlooked repertoire in the dynamic microRNAome.
Trends Genet. 28:544–549. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lee LW, Zhang S, Etheridge A, Ma L, Martin
D, Galas D and Wang K: Complexity of the microRNA repertoire
revealed by next generation sequencing. RNA. 16:2170–2180. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Guo L and Chen F: A Challenge for miRNA:
Multiple isomiRs in miRNAomics. Gene. 544:1–7. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Burroughs AM, Ando Y, de Hoon MJ, Tomaru
Y, Nishibu T, Ukekawa R, Funakoshi T, Kurokawa T, Suzuki H,
Hayashizaki Y and Daub CO: A comprehensive survey of 3′ animal
miRNA modification events and a possible role for 3′ adenylation in
modulating miRNA targeting effectiveness. Genome Res. 20:1398–1410.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Guo L, Li H, Lu J, Yang Q, Ge Q, Gu W, Bai
Y and Lu Z: Tracking miRNA precursor metabolic products and
processing sites through completely analyzing high-throughput
sequencing data. Mol Biol Rep. 39:2031–2038. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Loher P, Londin ER and Rigoutsos I: Isomir
expression profiles in human lymphoblastoid cell lines exhibit
population and gender dependencies. Oncotarget. 5:8790–8802. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Llorens F, Hummel M, Pantano L, Pastor X,
Vivancos A, Castillo E, Mattlin H, Ferrer A, Ingham M, Noguera M,
et al: Microarray and deep sequencing cross-platform analysis of
the mirRNome and isomiR variation in response to epidermal growth
factor. BMC Genomics. 14:3712013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Guo L, Yu J, Liang T and Zou Q:
miR-isomiRExp: A web-server for the analysis of expression of miRNA
at the miRNA/isomiR levels. Sci Rep. 6:237002016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Larkin MA, Blackshields G, Brown NP,
Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm
A, Lopez R, et al: Clustal W and clustal X version 2.0.
Bioinformatics. 23:2947–2948. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Tamura K, Stecher G, Peterson D, Filipski
A and Kumar S: MEGA6: Molecular evolutionary genetics analysis
version 6.0. Mol Biol Evol. 30:2725–2729. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Finn RD, Bateman A, Clements J, Coggill P,
Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J,
et al: Pfam: The protein families database. Nucleic Acids Res.
42:D222–D230. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Letunic I, Doerks T and Bork P: SMART:
Recent updates, new developments and status in 2015. Nucleic Acids
Res. 43:257–260. 2014. View Article : Google Scholar
|
|
39
|
Guo L, Yu J, Yu H, Zhao Y, Chen S, Xu C
and Chen F: Evolutionary and expression analysis of miR-#-5p and
miR-#-3p at the miRNAs/isomiRs levels. Biomed Res Int.
2015:1683582015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Guo L, Zhang H, Zhao Y, Yang S and Chen F:
Selected isomiR expression profiles via arm switching? Gene.
533:149–155. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Guo L, Zhao Y, Zhang H, Yang S and Chen F:
Close association between paralogous multiple isomiRs and
paralogous/orthologues miRNA sequences implicates dominant sequence
selection across various animal species. Gene. 527:624–629. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Guo L, Chen F and Lu Z: Multiple IsomiRs
and Diversity of miRNA Sequences Unveil Evolutionary Roles and
Functional Relationships Across Animals. MicroRNA and Non-Coding
RNA: Technology, Developments and Applications. 127–144. 2013.
|