Introduction
Hepatocellular carcinoma (HCC) is a malignant tumor
with one of the highest rates of mortality worldwide, and its
associated morbidity and mortality remain of significant concern.
In accordance with the World Health Organization GLOBOCAN database,
HCC was the sixth most common cancer in 2012 (782,000 new cancer
cases worldwide, 5.6% of the total) and the second major cause of
cancer death in 2012 (746,000 deaths, 9.1% of the total) (1).
Measuring levels of tumor biomarkers for HCC is an
important tool for disease management and treatment, as elucidating
factors that regulate cancer resistance and metastasis may aid in
the development of effective strategies to treat and potentially
cure cancer. A previous study established that the combination of
three tumor markers, α-fetoprotein (AFP), AFP-L3, and des-γ
carboxyprothrombin results in good predictive ability for patient
survival following diagnosis (2).
In addition, a recent study reported that Forkhead box M1
overexpression promotes HCC cell proliferation by cell cycle
regulation (3). Numerous factors
participate in liver carcinogenesis, including deregulation of
microRNAs (miRNAs) (4); for
example, miRNA-142-3p and miRNA-143-5p have been identified to be
downregulated in HCC and exhibit synergistic effects on cell
motility (5), with the epigenetic
profile also observed to be changed in human HCC (6). Global gene expression profiles aid in
the understanding of the transcriptomic landscape and molecular
mechanism of HCC. In order to systematically study the
transcriptomic differences between HCC and adjacent tissues, and
elucidate the molecular mechanisms underlying HCC, the Agilent
Whole Human Genome Oligo Microarray (4×44 K) was used, and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR) was
used to screen and confirm the differentially expressed genes
between HCC and normal adjacent tissues.
Tumor necrosis factor (TNF) receptor superfamily
member 12A (TNFRSF12A; also known as CD266, FN14 and TWEAKR), is
the smallest member of the TNF superfamily of receptors that lacks
the cytoplasmic death domain (7),
and has been reported to be elevated in various types of cancer,
including HCC. TNFRSF12A overexpression in HCC has additionally
been correlated with poor surgical outcome (8). TNFRSF12A has been previously
identified to be upregulated in alcoholic hepatitis and is
increased in experimental models of liver injury (9). It is additionally considered as a
factor that promotes prostate cancer bone metastasis (10) and ectopic expression of TNFRSF12A
increases the invasive activity of prostate cancer cells (11). In various cancer cell lines,
including gastric cancer cell lines, TNFRSF12A has been observed to
be upregulated during 5-fluorouracil (5-FU) treatment, which
promotes resistance to 5-FU (12).
Although TNFRSF12A has been reported to serve an important role in
the development of various types of cancer, progression and
resistance, howver the underlying molecular mechanisms in HCC
remain to be fully elucidated. In the current study, TNFRSF12A was
knocked down in the SMMC7721 cell line through siRNA. Cells in
which TNFRSF12A was knocked down exhibited reduced reproductive and
metastatic capacity ex vivo. These results were consistent
with previous reports, for example Zhou et al (13,14)
reported that immunotoxins targeting the TNFRSF12A receptor can
induce melanoma cell necrosis, suggesting that TNFRSF12A may be a
candidate therapeutic target for cancer including HCC.
Thus, the present study aids in the systematic
investigation of the molecular mechanism underlying HCC, and
additional genes which exhibit significantly different expression
levels between normal adjacent tissues and HCC tissues.
Materials and methods
Tissue specimens and cell lines
The current study was approved by the ethics
committee of Renji Hospital, Shanghai Jiao Tong University School
of Medicine (Shanghai, China). A total of 20 patients (12 male, 8
female) with confirmed case of liver disease form Renji Hospital
between July 2014 and July 2015 were included in the current study
and the average age was 58.8±1.2 years, the cancerous liver tissues
were obtained during surgery. Written informed consent was obtained
from each patient prior to the use of their tissue sample for
scientific research. Tumor and adjacent liver tissues from surgical
specimens were frozen in liquid nitrogen immediately until use. The
human HCC cell line, SMMC7721, was obtained from the American Type
Culture Collection (Manassas, VA, USA) and was cultured in complete
RPMI-1640 medium containing 10% fetal bovine serum (FBS;
No-Worries™; Thermo Fisher Scientific, Inc., Waltham, MA, USA) and
100 U/ml penicillin/streptomycin (Invitrogen; Thermo Fisher
Scientific, Inc.) at 37°C in a humidified atmosphere of 5%
CO2.
Microarray expression analysis
Total RNA from each sample was quantified using the
NanoDrop ND-1000 (Thermo Fisher Scientific, Inc.) and the RNA
integrity was assessed using standard denaturing agarose gel
electrophoresis. For microarray analysis, the Agilent array
platform was used. The sample preparation and microarray
hybridization were performed based on the manufacturer's standard
protocols. Briefly, total RNA from each sample was amplified and
transcribed into fluorescent cRNA with using Agilent Quick Amp
Labeling protocol (version 5.7; Agilent Technologies). The labeled
cRNAs were hybridized onto the Whole Human Genome Oligo Microarray
(4×44K; Agilent Technologies, Inc., Santa Clara, CA, USA).
Subsequent to washing of the slides with potassium permanganate ,
the arrays were scanned using the Agilent Scanner G2505C (Agilent
Technologies, Inc.).
Agilent Feature Extraction software (version
11.0.1.1; Agilent Technologies, Inc.) was used to analyze the
acquired array images. Quantile normalization and subsequent data
processing were performed using GeneSpring GX software, version
11.5.1 (Agilent Technologies, Inc.).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was isolated using TRIzol (Life
Technologies; Thermo Fisher Scientific, Inc.) according to the
manufacturer's instructions. Extracted RNA was quantitated using
NanoDrop ND-2000 (Thermo Fisher Scientific, Inc.) and treated with
DNase I (Life Technologies; Thermo Fisher Scientific). The reverse
transcription reaction was performed using PrimeScript™ RT Master
Mix (Takara Biotechnology Co., Ltd., Dalian, China). RT-qPCR was
performed on a Applied Biosystems 7500 machine using SYBR Green PCR
Master Mix (Life Technologies; Thermo Fisher Scientific, Inc.).
GAPDH was used as a loading control. The ∆∆Cq method was used to
calculate the fold change in the expression of each gene (15).
siRNA transfection in SMMC7721
cells
A non-targeting siRNA control [negative control
(NC); cat. no. D-001320-01-50] was purchased from ABgene (Thermo
Fisher Scientific, Inc.). TNFRSF12A-homo-400 (si-TNFRSF12A; cat.
no. Q000007132-1-A) was purchased from Guangzhou RiboBio Co., Ltd.
(Guangzhou, China). Cells were transfected using Lipofectamine 2000
in OptiMEM (Life Technologies; Thermo Fisher Scientific, Inc.). The
knockdown of TNFRSF12A was quantified 48 h post-transfection using
western blotting.
Western blot analysis
Western blot analysis was used to assess TNFRSF12A
protein reduction. Cells were harvested in RIPA lysis buffer
containing 50 mM Tris, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% Triton
N-100, 1% sodium deoxycholate and 0.1% sodium dodecyl sulfate and
protease inhibitor cocktail (Roche Diagnostics, Basel, Switzerland)
48 h after siRNA transfection. The protein concentration was
quantified using the Bio-Rad DC protein assay kit (Bio-Rad
Laboratories, Inc. Hercules, CA, USA). Equal amounts of protein
were separated using 12% SDS-PAGE and transferred to nitrocellulose
membranes. The membranes were blocked with 5% nonfat dried milk in
Tris-buffered saline, pH 8.0, with 0.1% Tween 20 for 1 h. The
primary antibodies used included rabbit anti-human TNFRSF12A
(Abcam, Cambridge, UK) and β-tubulin (R&D Systems, Inc.,
Minneapolis, MN, USA). Membranes were washed with SuperSignal West
dura (Thermo Fisher Scientific, Inc.) prior to visualizing the
protein bands under the imaging apparatus (GE Healthcare Life
Sciences, Chalfont, UK). Densitometric analysis was performed with
ImageJ version 2.1.4.7 (National Institutes of Health, Bethesda,
MD, USA).
Cell growth assay
Cells were plated into 96-well plates at a density
of 3×103 cells/well and were maintained at 37°C in a
humidified incubator. Subsequent to the initial transfection with
the different siRNAs, cell growth was measured at 24, 48 and 72 h
by adding 20 µl/well MTT (5 mg/ml; Sigma-Aldrich; Merck Millipore,
Darmstadt, Germany). Plates were then incubated for an additional 4
h at 37°C. Culture medium was removed from the wells, then dimethyl
sulfoxide (Sigma-Aldrich; Merck Millipore) was added at 100
µl/well. Plates were then incubated for 20 min in a 37°C incubator,
and absorbance was measured with the microplate reader (Bio-Rad
Laboratories, Inc.) at a wavelength of 495 nm. Each experiment was
repeated three separate times.
Transwell migration assay
Micrometer pore size translucent transwell migration
chambers (BD Biosciences, Franklin Lakes, NJ, USA) pre-coated with
50 µl Matrigel (50 µg Matrigel in Dulbecco's modified Eagle's
medium; Corning Incorporated, Corning, NY, USA) were plated in a
24-well plate. The lower chamber was filled with medium containing
10% FBS as a chemoattractant, and after transfection with the
different siRNAs for 24 h, 1×105 cells were transferred
to the upper chamber in 100 µl serum-free medium. Following
incubation for 24, 48 and 72 h at 37°C in an environment with 5%
CO2, each transwell migration chamber was removed and
0.1% crystal violet (Sigma-Aldrich; Merck Millipore) was used to
stain the cells that passed through the filter. Cells adhering to
the bottom of the chamber were counted under a microscope (Olympus
Corporation, Tokyo, Japan) in a minimum of three random fields. The
mean number of cells was obtained in each group.
Statistical analysis
Statistical analyses of the differences between
groups were performed using Student's t-test. SPSS version 18
(SPSS, Inc., Chicago, IL, USA) was used to perform statistical
analyses. P<0.05 was considered to indicate a statistically
significant difference.
Results
Heatmap and gene ontology (GO)
analysis between HCC and adjacent tissues
Subsequent to gene expression normalization, the
genes identified to be significantly different between HCC and
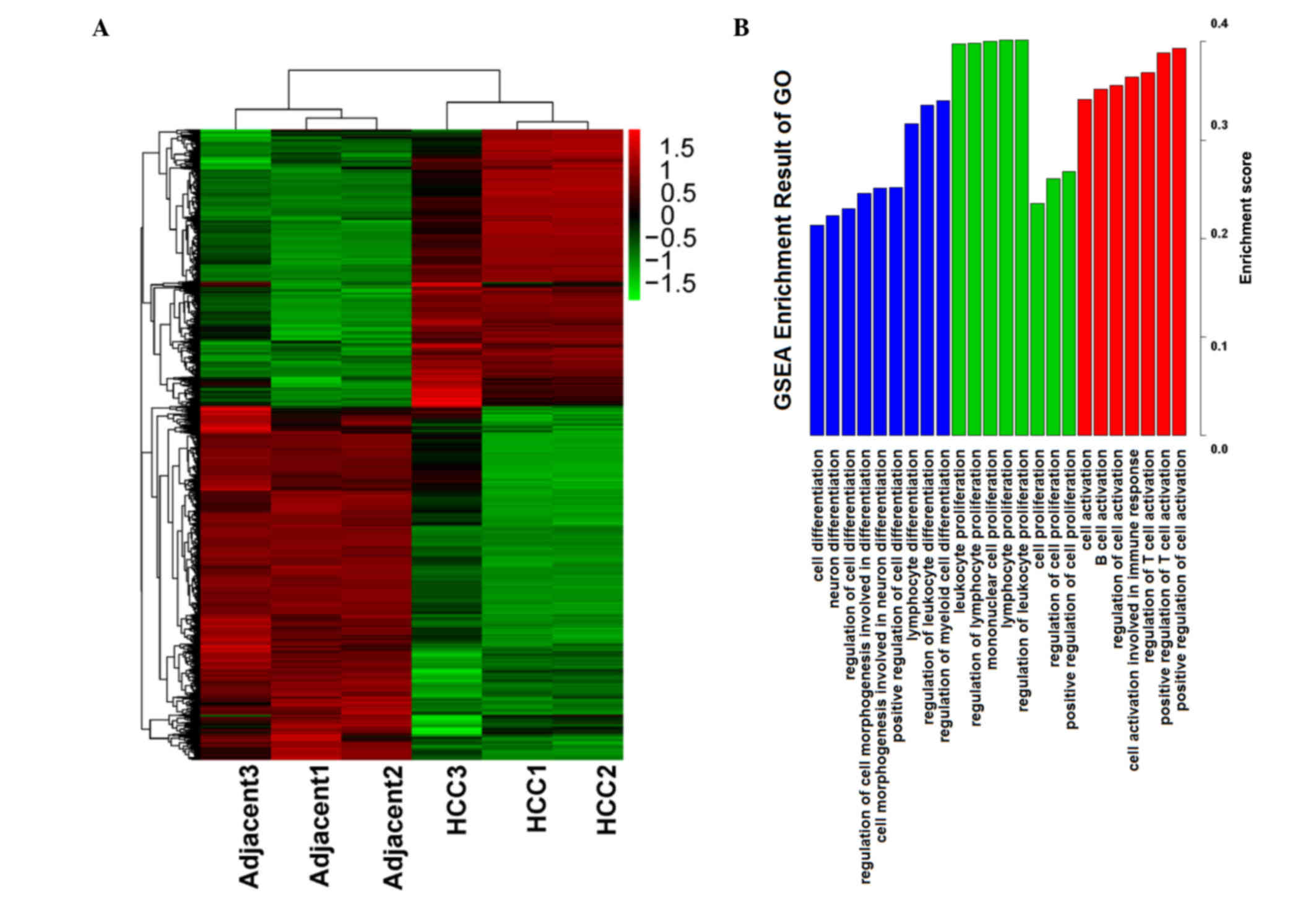
adjacent tissues by the t-test were selected, and a heatmap was
used to calculate a cluster result, (see Fig. 1A). Through the heatmap, the
difference of the two sample types was evident. The two types of
samples were separated into 2 clusters, and the differentially
expressed genes also generated 2 groups. These results indicate
that the gene expression data was reliable, thus the gene
expression data was used for the subsequent steps of analysis.
Which functions and features are involved in and
serve an important role in HCC is important, thus Gene Set
Enrichment Analysis (GSEA) was conducted, focussing on functions in
GO biological processes. Enrichment results from GSEA are presented
in Fig. 1B. Several main features
were selected in ordered by enrichment score. The main enrichment
results were the functions of activation, proliferation and
differentiation, and these functions predominantly serve a positive
role in regulation. Thus, it is suggested that in the process of
development and progression of cancer, proliferation and
differentiation serves an important role.
Numerous genes are significantly
different between HCC and adjacent tissues
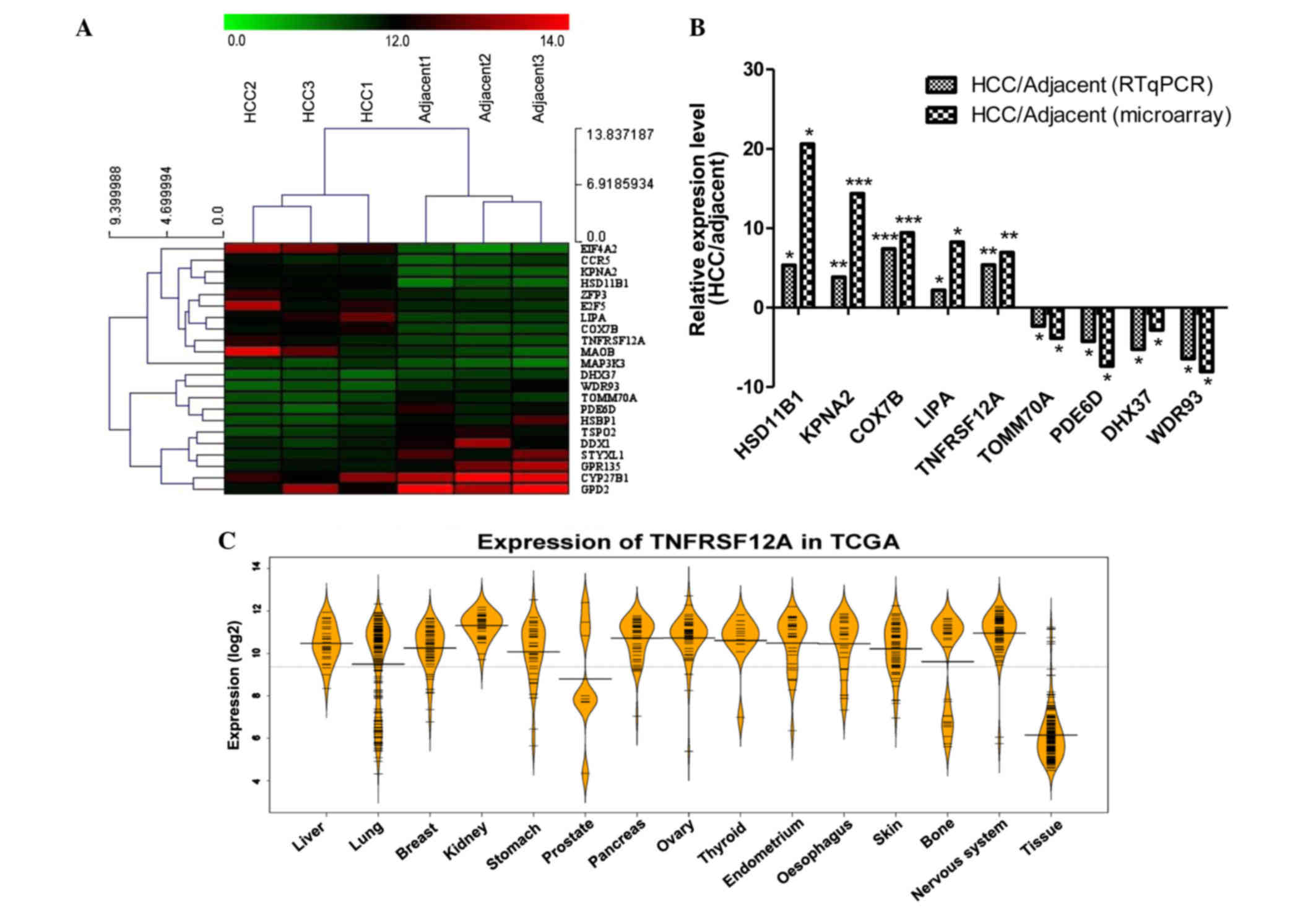
The top 22 differentially expressed genes were
selected from 6 samples, then MeV version 4.8 (mev.tm4.org/) was used for hierarchical clustering
(Fig. 2A). The cluster result
demonstrated that the 3 HCC samples and 3 adjacent samples are
divided into two different groups. This indicates that the
expression of these genes can separate the HCC and adjacent samples
by the hierarchical clustering method. In addition, the cluster
result for the genes also generated 2 groups, the expression of
genes of the first group in the HCC sample was greater than that of
the adjacent samples. The features of these 12 genes may serve a
specific role in HCC. The expression of additional genes was
greater in the adjacent samples than in the HCC samples (Table I). In addition, the expression
levels of cytochrome P450 family 27 subfamily B member 1 (CYP27B1)
and glycerol-3-phosphate dehydrogenase 2 (GPD2) were high in the
HCC and adjacent samples, suggesting that these two genes serve an
important role in the two sample types. The results were in
agreement with previous studies, for example, hydroxysteroid (11-β)
dehydrogenase 1 (HSD11B1), the top upregulated genes in the HCC
samples in the current study, was previously reported to be
upregulated in patients with liver cancer (16,17).
 | Table I.Gene expression profiling of HCC and
adjacent samples. |
Table I.
Gene expression profiling of HCC and
adjacent samples.
| Gene | HCC1 | HCC2 | HCC3 | Adjacent1 | Adjacent2 | Adjacent3 |
|---|
| HSD11B1 | 11.76468 | 11.3246 | 11.54463 | 6.037427 | 8.3245 | 7.180964 |
| KPNA2 | 11.226649 | 11.6355 | 11.43109 | 7.338309 | 7.8375 | 7.587905 |
| COX7B | 12.179636 | 11.3544 | 11.76702 | 8.827969 | 8.235 | 8.531484 |
| LIPA | 12.815084 | 11.6589 | 12.23699 | 9.059839 | 9.3255 | 9.19267 |
| ZFP3 | 10.863089 | 12.4356 | 11.64934 | 10.46896 | 9.3255 | 9.897231 |
| EIF4A2 | 12.342399 | 13.3479 | 12.98746 | 7.819575 | 5.2345 | 6.3456 |
| TNFRSF12A | 10.364899 | 12.3245 | 11.3447 | 8.957959 | 8.3456 | 8.34566 |
| MAP3K3 | 9.738604 | 9.23456 | 8.273484 | 8.09329 | 7.23455 | 6.238847 |
| MAOB | 10.040855 | 13.7844 | 12.76466 | 9.658285 | 8.93774 | 7.377461 |
| CCR5 | 10.378646 | 11.3765 | 10.34764 | 7.098104 | 8.38746 | 9.378645 |
| E2F5 | 12.297534 | 13.3746 | 11.32374 | 10.19454 | 10.34662 | 9.37462 |
| TOMM70A | 8.951378 | 8.3875 | 7.934875 | 11.35481 | 10.37468 | 9.376462 |
| PDE6D | 9.847308 | 8.23675 | 7.374662 | 12.33905 | 10.38642 | 11.36525 |
| DHX37 | 7.104071 | 7.23642 | 8.323786 | 8.647532 | 9.238765 | 9.236455 |
| WDR93 | 7.613555 | 7.23765 | 8.232386 | 9.800161 | 10.37452 | 11.93478 |
| HSBP1 | 9.246032 | 8.23885 | 8.23865 | 11.5736 | 10.37846 | 12.62356 |
| DDX1 | 10.776629 | 10.2387 | 9.237658 | 12.12334 | 13.23775 | 11.76325 |
| TSPO2 | 10.47408 | 8.32486 | 9.236746 | 11.80525 | 12.23877 | 11.23876 |
| GPD2 | 12.100197 | 11.239 | 13.2384 | 14.55473 | 13.39845 | 14.23987 |
| CYP27B1 | 13.075796 | 12.3896 | 11.93784 | 13.39113 | 14.23789 | 13.78467 |
| STYXL1 | 10.783392 | 9.98746 | 10.35623 | 12.57094 | 11.23898 | 12.87364 |
| GPR135 | 10.884162 | 10.3898 | 11.23876 | 11.55622 | 12.89348 | 13.38763 |
RT-qPCR was used to confirm the differences in gene
expression (Fig. 2B). Among the 22
genes analyzed, 9 genes which have not been previously investigated
were selected and were differently expressed in the microarray
confirming the RT-qPCR findings. Among them, cyclooxygenase 7B
(COX7B) has been reported to be overexpressed in nasopharyngeal
carcinoma in a similar manner (18), as a previously known
chemoresistance gene, COX7B also has been identified to be
upregulated in post-chemotherapeutic ovarian tumors (19). WD repeat domain 93 (WDR93) which
was identified to be downregulated in HCC tumor samples and was not
previously associated with disease, was identified to be associated
with neurological disorders through whole-exome sequencing
(20). In controlling neutral
lipid metabolism, liver homeostasis, immune response and tumor
metastasis, lipase [also termed LAL, encoded by lipase A, lysosomal
acid type (LIPA)] is a critical metabolic enzyme, which was
identified in the current study to be upregulated. Tumor-promoting
myeloid-derived suppressive cells in the liver of LAL−/−
mice were reported to be reduced by human LIPA expression (21). TNFRSF12A expression in HCC tissue
was detected to be 5.3 and 6.9 times higher than in adjacent tissue
through RT-qPCR and microarray, respectively.
To illustrate the difference of TNFRSF12A gene
expression levels between cancer and normal tissue, The Cancer
Genome Atlas (TCGA) was used. TCGA is a comprehensive effort to
accelerate understanding of the molecular basis of cancer through
the use of genome analysis technologies, including large-scale
genomic sequencing. Using TCGA, 3-level expression data was
downloaded; the data contained 14 cancer sample types and 1 normal
tissue sample type. The expression of TNFRSF12A in the cancer
sample type was in general greater than that of the normal sample
type (Fig. 2C), which was
consistent with previous studies.
TNFRSF12A knockdown reduced SMMC7721
cell viability
TNFRSF12A, the smallest member of the TNF
superfamily of receptors that lacks the cytoplasmic death domain
(7), has been reported to be
elevated in various types of cancer, including HCC, which was
confirmed by clear upregulation in HCC tissues via microarray and
RT-qPCR (Fig. 2 and Table I). In order to investigate the
underlying molecular mechanisms in HCC, TNFRSF12A was selected for
knockdown in the SMMC7721 cell line through siRNA. A total of 48 h
after initial transfection, TNFRSF12A was detected to be
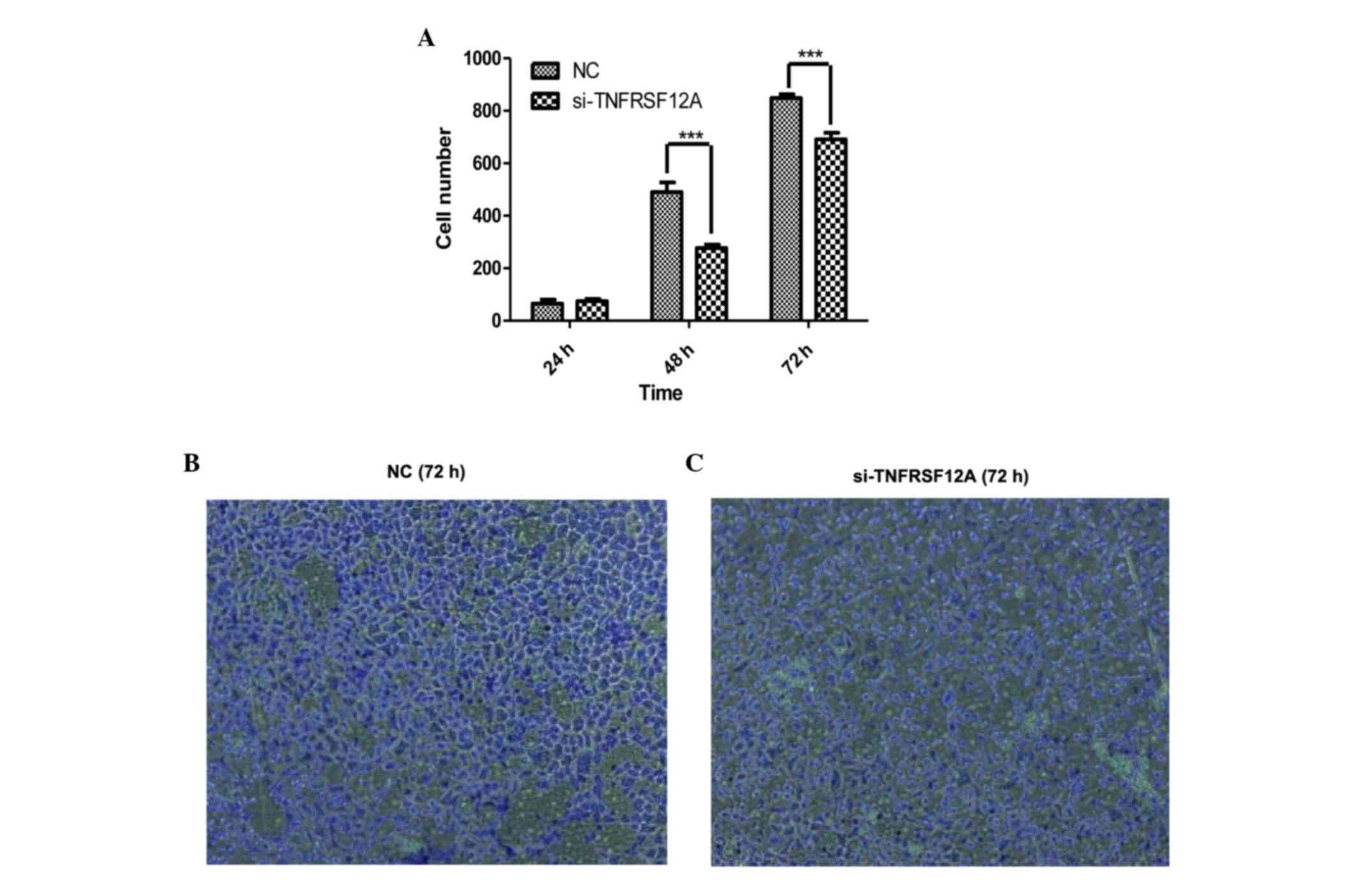
downregulated through western blotting (Fig. 3A and B). Cell growth was measured
24, 48 and 72 h after initial siRNA transfection and according to
the cell growth curves (Fig. 3C),
cell viability was stronger in the si-TNFRSF12A transfected group
in comparison with that of the NC-siRNA transfected group,
suggesting that TNFRSF12A increases SMMC7721 cell viability in
vitro.
TNFRSF12A knockdown can reduce
invasive potential and metastatic capacity of SMMC7721 cells in
vitro
Acting via cell-secreted proteolytic degradation of
the cellular basement membrane, tumor cells invasion is the leading
cause of cancer-associated mortality (22). Compared with the NC group, SMMC7721
cells which were transfected with si-TNFSFR12A exhibited greatly
inhibited invasive potential and metastatic capacity. The number of
cells passing through the membrane was significantly reduced in the
si-TNFSRF12A group at 48 and 72 h (Fig. 4).
Discussion
HCC is a tumor type that is highly insensitive to
conventional chemotherapy (23),
and increasingly targeted molecular therapies have exhibited
significant benefits in patients with cancer, including those with
HCC. The eludication of the molecular mechanism of HCC occurrence
and development is important for the development of effective
treatments for HCC. Thus, in the current study, the Agilent Whole
Human Genome Oligo Microarray (4×44 K) was used to investigate the
differentially expressed genes between HCC and adjacent tissues,
and the top 22 differentially expressed genes were confirmed by
RT-qPCR. Among these large differences in gene expression,
TNFRSF12A expression in HCC tissue was identified to be greater
than that of adjacent tissue. The TNFRSF12A receptor has been
previously identified to promote the invasive potential and
metastatic capacity of non-small lung adenocarcinoma cells through
the upregulation of integrin α6 (24). As the sole signaling receptor for
the proinflammatory cytokine TWEAK (TNFSF12), TNFRSF12A engagement
stimulates multiple signal transduction pathways, including the
nuclear factor κB pathway, and it has been previously identified
that TNFRSF12A may serve a role in tumor growth and metastasis
(25). in the current study,
TNFRSF12A was knocked down in a SMMC7721 cell line through siRNA,
and cells exhibited reduced reproductive and metastatic capacity
ex vivo. Thus, the present study suggests that TNFRSF12A may
be a candidate therapeutic target for cancer including HCC, and
significant differentially expressed genes between HCC and normal
adjacent tissues require further investigation in subsequent
studies.
References
|
1
|
Deng GL, Zeng S and Shen H: Chemotherapy
and target therapy for hepatocellular carcinoma: New advances and
challenges. World J Hepatol. 7:787–798. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Toyoda H, Kumada T, Tada T, Sone Y,
Kaneoka Y and Maeda A: Tumor markers for hepatocellular carcinoma:
Simple and significant predictors of outcome in patients with HCC.
Liver Cancer. 4:126–136. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yu M, Tang Z, Meng F, Tai M, Zhang J, Wang
R, Liu C and Wu Q: Elevated expression of FoxM1 promotes the tumor
cell proliferation in hepatocellular carcinoma. Tumour Biol.
37:1289–1297. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wang Y and Tian Y: miRNA for diagnosis and
clinical implications of human hepatocellular carcinoma. Hepatol
Res. 46:89–99. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Tsang FH, Au SL, Wei L, Fan DN, Lee JM,
Wong CC, Ng IO and Wong CM: MicroRNA-142-3p and microRNA-142-5p are
downregulated in hepatocellular carcinoma and exhibit synergistic
effects on cell motility. Front Med. 9:331–343. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Nishida N and Kudo M: Alteration of
epigenetic profile in human hepatocellular carcinoma and its
clinical implications. Liver Cancer. 3:417–427. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wiley SR, Cassiano L, Lofton T,
Davis-Smith T, Winkles JA, Lindner V, Liu H, Daniel TO, Smith CA
and Fanslow WC: A novel TNF receptor family member binds TWEAK and
is implicated in angiogenesis. Immunity. 15:837–846. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li N, Hu WJ, Shi J, Xue J, Guo WX, Zhang
Y, Guan DX, Liu SP, Cheng YQ, Wu MC, et al: Roles of fibroblast
growth factor-inducible 14 in hepatocellular carcinoma. Asian Pac J
Cancer Prev. 14:3509–3514. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Affò S, Dominguez M, Lozano JJ, Sancho-Bru
P, Rodrigo-Torres D, Morales-Ibanez O, Moreno M, Millán C,
Loaeza-del-Castillo A, Altamirano J, et al: Transcriptome analysis
identifies TNF superfamily receptors as potential therapeutic
targets in alcoholic hepatitis. Gut. 62:452–460. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yin J, Liu YN, Tillman H, Barrett B,
Hewitt S, Ylaya K, Fang L, Lake R, Corey E, Morrissey C, et al:
AR-regulated TWEAK-FN14 pathway promotes prostate cancer bone
metastasis. Cancer Res. 74:4306–4317. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Huang M, Narita S, Tsuchiya N, Ma Z,
Numakura K, Obara T, Tsuruta H, Saito M, Inoue T, Horikawa Y, et
al: Overexpression of Fn14 promotes androgen-independent prostate
cancer progression through MMP-9 and correlates with poor treatment
outcome. Carcinogenesis. 32:1589–1596. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kwon OH, Kim JH, Kim SY and Kim YS:
TWEAK/Fn14 signaling mediates gastric cancer cell resistance to
5-fluorouracil via NF-kB activation. Int J Oncol. 44:583–590.
2014.PubMed/NCBI
|
|
13
|
Zhou H, Hittelman WN, Yagita H, Cheung LH,
Martin SS, Winkles JA and Rosenblum MG: Antitumor activity of a
humanized, bivalent immunotoxin targeting fn14-positive solid
tumors. Cancer Res. 73:4439–4450. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhou H, Ekmekcioglu S, Marks JW,
Mohamedali KA, Asrani K, Phillips KK, Brown SA, Cheng E, Weiss MB,
Hittelman WN, et al: The TWEAK receptor Fn14 is a therapeutic
target in melanoma: Immunotoxins targeting Fn14 receptor for
malignant melanoma treatment. J Invest Dermatol. 133:1052–1062.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Pan ZQ, Fang ZQ and Lu WL: Characteristics
of gene expression of adrenal cortical steroid synthetase and its
regulatory factor in mice with H22 liver cancer of different
patterns. Zhongguo Zhong Xi Yi Jie He Za Zhi. 31:85–89. 2011.(In
Chinese). PubMed/NCBI
|
|
17
|
Soldan M, Nagel G, Losekam M, Ernst M and
Maser E: Interindividual variability in the expression and NNK
carbonyl reductase activity of 11beta-hydroxysteroid dehydrogenase
1 in human lung. Cancer Lett. 145:49–56. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Xiong S, Wang Q, Zheng L, Gao F and Li J:
Identification of candidate molecular markers of nasopharyngeal
carcinoma by tissue microarray and in situ hybridization. Med
Oncol. 28:(Suppl 1). S341–S348. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
L'Espérance S, Popa I, Bachvarova M,
Plante M, Patten N, Wu L, Têtu B and Bachvarov D: Gene expression
profiling of paired ovarian tumors obtained prior to and following
adjuvant chemotherapy: Molecular signatures of chemoresistant
tumors. Int J Oncol. 29:5–24. 2006.PubMed/NCBI
|
|
20
|
Alazami AM, Patel N, Shamseldin HE, Anazi
S, Al-Dosari MS, Alzahrani F, Hijazi H, Alshammari M, Aldahmesh MA,
Salih MA, et al: Accelerating novel candidate gene discovery in
neurogenetic disorders via whole-exome sequencing of prescreened
multiplex consanguineous families. Cell Rep. 10:148–161. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Du H, Zhao T, Ding X and Yan C:
Hepatocyte-Specific expression of human lysosome acid lipase
corrects liver inflammation and tumor metastasis in lal(−/−) mice.
Am J Pathol. 185:2379–2389. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Peng HX, Wu WQ, Yang DM, Jing R, Li J,
Zhou FL, Jin YF, Wang SY and Chu YM: Role of B7-H4 siRNA in
proliferation, migration, and invasion of LOVO colorectal carcinoma
cell line. Biomed Res Int. 2015:3269812015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Liang XD, Dai YC, Li ZY, Gan MF, Zhang SR,
Yin-Pan Lu HS, Cao XQ, Zheng BJ, Bao LF, et al: Expression and
function analysis of mitotic checkpoint genes identifies TTK as a
potential therapeutic target for human hepatocellular carcinoma.
PLoS One. 9:e977392014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Jandova J, Mason CJ, Pawar SC and Watts
GS: Fn14 receptor promotes invasive potential and metastatic
capacity of non-small lung adenocarcinoma cells through the
up-regulation of integrin α6. Neoplasma. 62:41–52. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Cheng E, Whitsett TG, Tran NL and Winkles
JA: The TWEAK receptor Fn14 Is an Src-inducible protein and a
positive regulator of Src-driven cell invasion. Mol Cancer Res.
13:575–583. 2015. View Article : Google Scholar : PubMed/NCBI
|