Introduction
Osteosarcoma is characterized as a primary bone
malignancy, which originates from bone-forming mesenchymal cells
and commonly occurs in children and young adults, with a second
incidence peak occurring in adulthood (1,2). The
pulmonary metastasis of osteosarcoma is responsible for the
majority of cases of mortality in patients, due to the lack of
effective therapeutic methods (3).
Previous studies have aimed to develop specific therapeutic
approaches targeting metastatic progression, and to understand the
mechanisms underlying metastasis (4,5).
Cysteine-rich angiogenic inducer 61 has been reported to promote
the epithelial to mesenchymal transition of osteosarcoma (6). In addition, Ezrin expression has been
reported to be necessary for metastasis of osteosarcoma (7), and high expression levels of
transforming growth factor-β (TGF-β) have been observed in
osteosarcoma cell lines (8).
However, these previous findings are insufficient and gene
expression profiling is required to comprehensively analyze the key
genes associated with osteosarcoma metastasis.
Transcription factors (TFs) (9) and microRNAs (miRNAs) (10) are the two critical regulators
targeting protein-coding genes involved in cancer initiation and
progression. For example, runt-related transcription factor 2 may
support the cell adhesion and motility of osteosarcoma cells
(11). In addition, it has
previously been reported that overexpression of SRY (sex
determining region Y)-box 9, Wnt family member 1 and frizzled class
receptor 1 may be associated with an advanced clinical stage
(12), whereas inhibition of Wnt
may markedly reduce the lung metastasis of osteosarcoma (13). miRNAs also contribute to
osteosarcoma and have potential applications as therapeutic targets
(14). For example, miRNA
(miR)-143 downregulation may result in matrix metalloproteinase 13
upregulation, and thus promote lung metastasis of human
osteosarcoma (15). Furthermore,
miR-199a-3p and miR-34a may inhibit osteosarcoma cell growth and
migration via downregulation of target genes, such as Met,
mTOR and Stat3 (16,17).
Therefore, the identification of these TFs and miRNAs may aid in
the determination of regulatory network alterations over the
progression of osteosarcoma.
The present study aimed to improve the understanding
of the molecular mechanisms underlying osteosarcoma metastasis. The
differentially expressed genes (DEGs) between metastatic
osteosarcoma and non-metastatic osteosarcoma cells were determined
by analyzing the GSE49003 public dataset deposited in the Gene
Expression Omnibus (GEO), providers of which have not used the data
to publish an article at present. Subsequently, identification of
the associated TFs and miRNAs of DEGs was conducted using
bioinformatics methods. Enrichment analysis was also performed on
the DEGs to determine the metastasis-associated molecular
mechanisms.
Materials and methods
Microarray data
Microarray data with accession number GSE49003 was
downloaded from the GEO database (http://www.ncbi.nlm.nih.gov/geo/) (18). This dataset contained data obtained
from KHOS and KRIB metastatic osteosarcoma cell lines, and HOS and
U2OS non-metastatic osteosarcoma cell lines. Each of the four cell
lines was analyzed in triplicate using Illumina HumanHT-12 V3.0
expression BeadChip (GPL6947; Illumina, Inc., San Diego, CA,
USA).
Data processing
The downloaded microarray data were processed
through missing value filtration with the criterion of probe
intensity values P>0.05 in >3 of the 12 expression profiles.
Kernel and nearest neighbor averaging methods (19) were used to impute the missing
values using the impute package (20) in R (version 3.13; http://www.r-project.org/). Quantile normalization was
performed to obtain standardized microarray data using
preprocessCore package (21) in R.
The probes were transformed into gene symbols based on the
annotation information of platform GPL6947. Subsequently, the gene
expression level of each gene was calculated by determining the
average of the expression levels of probes corresponding to the
same gene using aggregate package in R (22).
Screening of DEGs
In order to account for the innate differences among
various cell lines, one-way analysis of variance was performed to
exclude DEGs with higher significance in the intra-groups compared
with the inter-groups using the genefilter package (23) in R. Student's t-test was performed
to identify differential expression between metastatic and
non-metastatic cell lines using the limma package (24) in R. The Benjamini and Hochberg
method was applied to adjust the P-value with false discovery rate
(25). The DEGs were identified
under the thresholds of |log2 fold change (FC)|>1(FC
magnitude >2) and adjusted P<0.05.
Functional enrichment analysis
In order to determine the biological processes and
pathways associated with the DEGs, the online tool TargetMine
(http://targetmine.nibio.go.jp) (26) was used to perform Gene Ontology
(GO) function enrichment analysis based on GO (http://www.geneontology.org/) and UniProtKB GOA
(http://www.ebi.ac.uk/GOA) databases; pathway
enrichment analysis based on Kyoto Encyclopedia of Genes and
Genomes (http://www.genome.jp/kegg/) and
Reactome (http://www.reactome.org/) databases;
and Disease Ontology (http://disease-ontology.org/) enrichment analysis.
Construction of integrated regulatory
networks
In order to determine the regulatory networks of
DEGs, Target Mine (26) was used
to predict the TFs from the DEGs based on OregAnno (http://nar.oxfordjournals.org/content/early/2015/11/16/nar.gkv1203.abstract)
and AMADEUS (http://acgt.cs.tau.ac.il/amadeus/) databases; and
upstream miRNAs of DEGs based on miRBase (http://www.mirbase.org) and miRTarBase (http://mirtarbase.mbc.nctu.edu.tw/) databases.
Construction of the integrated regulatory network between TFs and
target genes, and the network between miRNAs and DEGs, was
conducted using Cytoscape software (version 3.0.0) (27).
Results
Screened DEGs
Under the thresholds of |log2 FC|>1
and adjusted P<0.05, a total of 456 DEGs were identified in the
metastatic osteosarcoma cell lines in comparison with the
non-metastatic osteosarcoma cell lines, consisting of 248
upregulated and 208 downregulated genes.
Significantly enriched terms
The upregulated genes were significantly enriched in
various pathways, including downregulation of TGF-β receptor
signaling and TGF-β receptor signaling activates SMADs; genes
enriched in these pathways included protein phosphatase 1,
regulatory subunit 15A (PPP1R15A), transforming growth
factor, β receptor II (TGFBR2) and ubiquitin
carboxyl-terminal hydrolase L5 (UCHL5). The upregulated
genes were also enriched in various biological processes and
different diseases, including Barrett's esophagus, esophageal
disease and lung cancer; genes enriched in these disease ontology
terms included epidermal growth factor receptor (EGFR),
insulin-like growth factor 2 mRNA binding protein 3
(IGF2BP3), runt-related transcription factor 3
(RUNX3) and secreted frizzled-related protein 1
(SFRP1). The top 5 significantly enriched terms are
presented in Table I.
 | Table I.Significantly enriched terms of
upregulated genes. |
Table I.
Significantly enriched terms of
upregulated genes.
| A, Pathways |
|---|
|
|---|
| Enriched term | P-value | Count | Genes |
|---|
| Interconversion of
polyamines (REACT_14805) |
3.94×10−4 | 2 | SAT1,
SMOX |
| Downregulation of
TGF-β receptor signaling [REACT_120727] |
3.21×10−3 | 3 | PPP1R15A,
TGFBR2, UCHL5 |
| TGF-β receptor
signaling activates SMADs [REACT_120850] |
5.83×10−3 | 3 | PPP1R15A,
TGFBR2, UCHL5 |
| Heme biosynthesis
[REACT_9465] |
6.81×10−3 | 2 | COX15,
UROD |
| Disease
[REACT_116125] |
7.11×10−3 | 24 | ALDOC, ANTXR2,
APOBEC3G, CYBA, EGFR, FRAT2, HEY1, IDS, ITPR3, LRP5, PFKP,
PPP1R15A, PRSS3, RPL22, RPL26L1, RPL5, RPS7, SFRP1, SH3KBP1,
SLC2A1, STX1A, TBL1X, TGFBR2, UCHL5 |
|
| B, Gene Ontology:
Biological process |
|
| Enriched term | P-value | Count | Genes |
|
| Cellular response
to zinc ion [GO:0071294] |
3.46×10−4 | 3 | MT1G, MT1M,
MT2A |
| Regulation of
insulin secretion [GO:0050796] |
1.17×10−3 | 7 | FOXA2, GLUD1,
ITPR3, LRP5, SFRP1, SLC2A1, STX1A |
| Response to zinc
ion [GO:0010043] |
1.25×10−3 | 3 | MT1G, MT1M,
MT2A |
| Dephosphorylation
of RNA polymerase II C-terminal domain [GO:0070940] |
1.26×10−3 | 2 | RPRD1A,
SSU72 |
| Regulation of
hormone secretion [GO:0046883] |
1.45×10−3 | 8 | FOXA2, GLUD1,
IL11, ITPR3, LRP5, SFRP1, SLC2A1, STX1A |
|
| C, Disease ontology
terms |
|
| Enriched term | P-value | Count | Genes |
|
| Osteosclerosis
[DOID:4254] |
1.17×10−3 | 3 | FAM20C, IL11,
LRP5 |
| Barrett's esophagus
[DOID:9206] |
1.67×10−3 | 4 | EGFR, IGF2BP3,
RUNX3, SFRP1 |
| Esophageal disease
[DOID:6050] |
8.31×10−3 | 4 | EGFR, IGF2BP3,
RUNX3, SFRP1 |
| Lung cancer
[DOID:1324] |
9.43×10−3 | 13 | ANPEP, BNIP3,
CDCP1, CYP27B1, EGFR, FRMD3, GCLM, IGF2BP3, PLAU, RUNX3, SAT1,
SFRP1, SLC2A1 |
| Endometrial cancer
[DOID:1380] |
1.02×10−2 | 7 | CYP27B1, EGFR,
ETV5, IGF2BP3, MUTYH, RUNX3, SLC2A1 |
The downregulated genes were significantly enriched
in various pathways, including extracellular matrix organization,
and collagen biosynthesis and modifying enzymes; genes enriched in
these pathways included collagen, type XII, α 1 (COL12A1),
collagen, type I, α 1 (COL1A1), collagen, type IV, α
1 (COL4A1) and collagen, type V, α 1 (COL5A1).
Downreglated genes were also enriched in the TGF-β signaling
pathway, including bone morphogenetic protein 4 (BMP4),
inhibitor of DNA binding 3 (ID3) and SMAD family member 6
(SMAD6), as well as in biological processes, such as
extracellular matrix organization and extracellular structure
organization (e.g. COL12A1, COL1A1, COL4A1 and
COL5A1). The top 5 significantly enriched terms are
presented in Table II.
 | Table II.Significantly enriched terms of
downregulated genes. |
Table II.
Significantly enriched terms of
downregulated genes.
| A, Pathways |
|---|
|
|---|
| Enriched term | P-value | Count | Genes |
|---|
| Extracellular
matrix organization [REACT_118779] |
3.29×10−5 | 13 | BMP4, CASK,
COL12A1, COL1A1, COL4A1, COL5A1, FBLN1, FBN2, PLOD2, SDC4,
SERPINH1, THBS1, TIMP2 |
| Collagen
biosynthesis and modifying enzymes [REACT_121139] |
1.77×10−4 | 6 | COL12A1, COL1A1,
COL4A1, COL5A1, PLOD2, SERPINH1 |
| Syndecan-1-mediated
signaling events [syndecan_1_pathway] |
3.12×10−4 | 5 | CASK, COL12A1,
COL1A1, COL4A1, COL5A1 |
| TGF-β signaling
pathway [hsa04350] |
5.99×10−4 | 6 | BMP4, FST, ID3,
PPP2CB, SMAD6, THBS1 |
| Legionellosis
[hsa05134] |
7.22×10−3 | 5 | CXCL8, EEF1A2,
HSPA2, IL18, NFKBIA |
|
| B, Gene ontology:
Biological process |
|
| Enriched term | P-value | Count | Genes |
|
| Extracellular
matrix organization [GO:0030198] |
4.94×10−6 | 15 | APP, BMP4, CASK,
COL12A1, COL1A1, COL4A1, COL5A1, FBLN1, FBN2, GAS6, PLOD2, SDC4,
SERPINH1, THBS1, TIMP2 |
| Extracellular
structure organization [GO:0043062] |
5.15×10−6 | 15 | APP, BMP4, CASK,
COL12A1, COL1A1, COL4A1, COL5A1, FBLN1, FBN2, GAS6, PLOD2, SDC4,
SERPINH1, THBS1, TIMP2 |
| Positive regulation
of cellular component movement [GO:0051272] |
6.48×10−6 | 13 | BMP4, COL1A1,
CXCL16, CXCL8, DAB2, GAS6, GLIPR2, GPER1, HSPA5, PDPN, SCARB1,
SEMA4D, THBS1 |
| Multicellular
organism development [GO:0007275] |
1.55×10−5 | 54 | ABLIM1, APP,
ARID5B, AXIN2, BASP1, BMP4, CACNA1H, CBX2, COL12A1, COL1A1, COL4A1,
COL5A1, CRABP2, CTGF, CXCL8, DAB2, DPYSL2, DUSP1, EBP, EPB41L3,
EYA2, FBN2, FES, FLOT2, FST, GAS6, GLIPR2, GPER1, HOXC6, HSPA5,
ID3, IGSF3, IL18, JUP, MDK, NDN, NINJ1, NPY, PDPN, PIM1, PLXNB2,
PQBP1, PRKCH, RRAS, SEMA4D, SFN, SHROOM3, SMAD6, TAGLN, THBS1,
TPM1, TRO, TUBB2B, VASN |
| Regulation of
cellular component movement [GO:0051270] |
1.98×10−5 | 18 | BMP4, COL1A1,
CXCL16, CXCL8, DAB2, FES, GAS6, GLIPR2, GPER1, HSPA5, JUP, PDPN,
PLXNB2, RCC2, SCARB1, SEMA4D, THBS1, TPM1 |
|
| C, Disease ontology
terms |
|
| Enriched term | P-value | Count | Genes |
|
| Embryoma
[DOID:4766] |
4.67×10−5 | 22 | AMPH, BMP4,
CALD1, CHD4, CTGF, CXCL8, DAB2, E2F2, GSTT1, HSPA5, IL18, KRT8,
MDK, NUAK1, PDPN, RBP1, SEMA4D, SHC1, SKP2, TIMP2, TK1,
TRO |
| Embryonal cancer
[DOID:688] |
5.20×10−5 | 22 | AMPH, BMP4,
CALD1, CHD4, CTGF, CXCL8, DAB2, E2F2, GSTT1, HSPA5, IL18, KRT8,
MDK, NUAK1, PDPN, RBP1, SEMA4D, SHC1, SKP2, TIMP2, TK1,
TRO |
| Cell type cancer
[DOID:0050687] |
6.41×10−5 | 33 | AMPH, APP, BMP4,
CALD1, CCNA2, CHD4, CTGF, CXCL8, DAB2, E2F2, FES, FST, GAS6, GSTT1,
HSPA5, ID3, IL18, IRF8, JUP, KRT8, MAGED1, MDK, NUAK1, PDPN, PLAT,
RBP1, SEMA4D, SHC1, SKP2, STIP1, TIMP2, TK1, TRO |
| Reproductive organ
cancer [DOID:193] |
1.27×10−4 | 24 | BMP4, C19orf33,
CACNA1H, CXCL16, CXCL8, DAB2, DUSP1, FST, FSTL1, GSTT1, HOXC8,
HSPA5, IL11RA, IL18, JUND, JUP, MDK, SEMA4D, SERPINH1, SHC1, SKP2,
TIMP2, TK1, TMSB15A |
| Liver disease
[DOID:409] |
1.41×10−4 | 9 | ALPP, CTGF,
CXCL16, CXCL8, GSTT1, IL18, KRT8, PLAT, RBP1 |
Analysis of regulatory networks
Based on the regulatory network between TFs and
target genes, 7 TFs targeting 30 genes were screened from the
upregulated genes, including FOS-like antigen 1 (FOSL),
forkhead box A2 (FOXA2) and peroxisome
proliferator-activated receptor γ (PPARG). In addition, 3
TFs targeting 19 genes were obtained from the downregulated genes,
including jun D proto-oncogene (JUND). Notably, PPARG
and JUND had a regulatory effect on the expression of
downstream spermidine/spermine N1-acetyltransferase 1 (SAT1)
(Fig. 1).
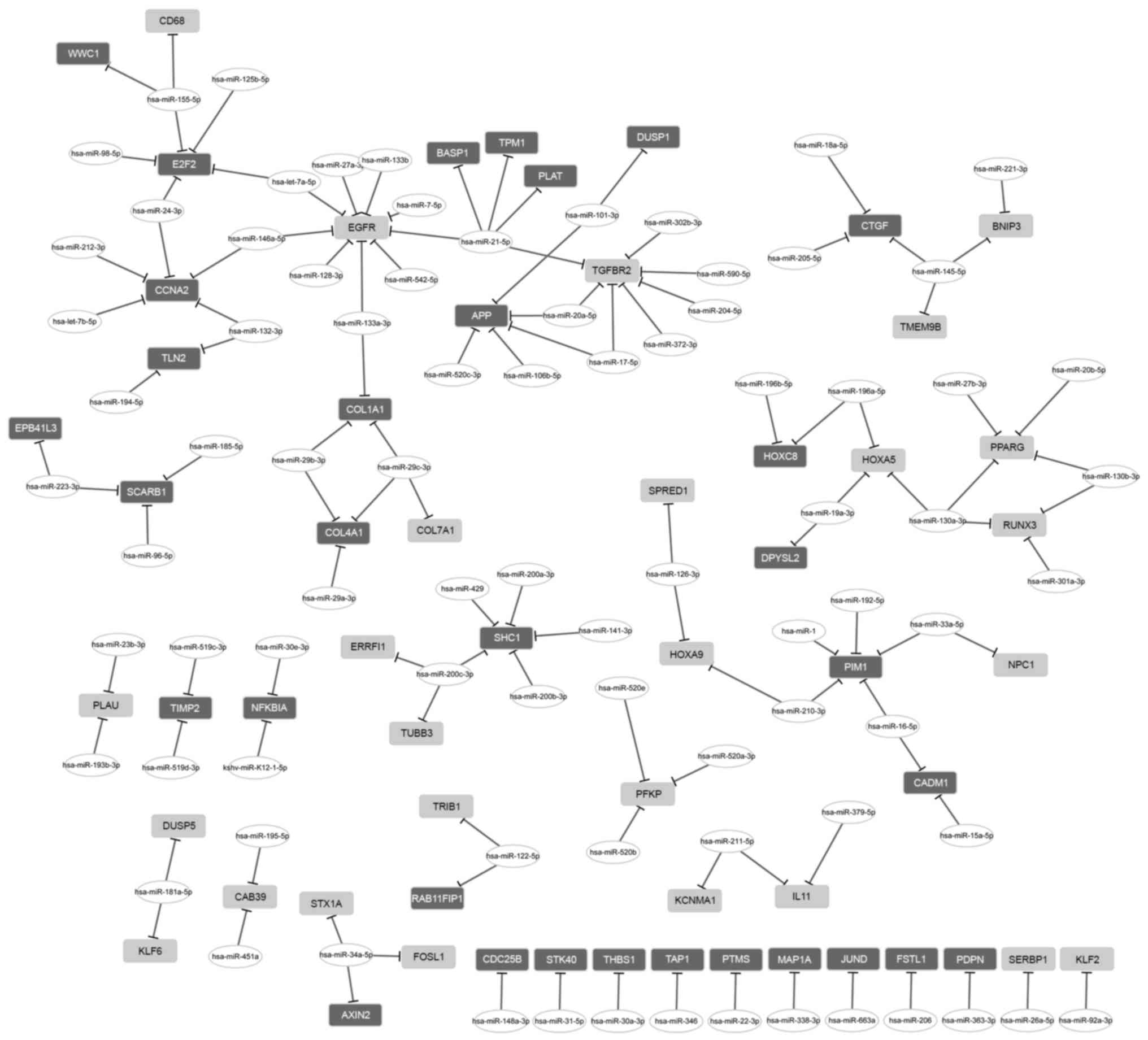
The regulatory network between miRNA and DEGs
consisted of 26 upregulated genes, 32 downregulated genes and 84
miRNAs (Fig. 2). From that
network, two miRNAs with a high degree were further investigated.
miR-21-5p had a regulatory effect on 5 DEGs, including tropomyosin
1 (TPM1), EGFR and TGFBR2; and miR-155-5p
regulated 3 DEGs, including E2F transcription factor 2
(E2F2).
Discussion
The present study presented similar research type to
a related study (28). The
similarities between the two studies were that the present study
used the same microarray data of GSE49003 to identify the DEGs in
the metastatic osteosarcoma samples and analyze the DEGs enriched
function and pathways, and the miRNA-target network. Additionally,
different results were obtained which may be due to the different
analytical tools used. The present study identified 248 upregulated
and 208 downregulated DEGs in metastatic cell lines compared with
in non-metastatic cell lines by analyzing a dataset deposited in
the GEO, in order to determine the molecular mechanisms underlying
osteosarcoma metastasis. The dysregulated genes were enriched in
TGF-β signaling, downregulation of TGF-β receptor signaling and
TGF-β receptor signaling activates SMADs pathways, as well as in
the lung cancer disease ontology term. A cluster of TFs and miRNAs
were also identified to be associated with the DEGs.
Pathway enrichment analysis revealed that the
upregulated PPP1R15A, TGFBR2 and UCHL5 genes
were significantly associated with the downregulation of TGF-β
receptor signaling and TGF-β receptor signaling activates SMADs
pathways, whereas the downregulated BMP4, ID3 and
SMAD6 were associated with the TGF-β signaling pathway.
Altered TGF-β expression was previously observed in patients
with metastatic osteosarcoma compared with in patients without
metastasis (29). In addition,
TGF-β may increase the metastatic potential of human osteosarcoma
cells by triggering the malignant phenotype presentation of
hyaluronan and versican (30).
Overexpressed SMAD7 may have an inhibitory effect on the
TGF-β/SMAD signaling pathway and subsequently protect against tumor
metastasis of osteosarcoma (31).
PPP1R15A and UCHL5 have previously been implicated in
the control of TGF-β signaling via interaction with SMAD7
(32,33). Furthermore, the TGF-β growth
factor family member BMP4 may suppress breast cancer
metastasis (34). ID genes
are the downstream targets of the TGF-β pathway, which contribute
to cell migration (35).
SMAD6, instead of SMAD7, may negatively regulate
TGF-β-induced activation of the TRAF6-TAK1-P38 MAPK/JNK pathway
(36). Therefore, it is possible
that TGF-β and TGF-β receptor signaling may contribute to
osteosarcoma metastasis, which may be mediated by the dysregulated
genes enriched in these pathways.
Disease ontology enrichment analysis revealed that
the upregulated DEGs EGFR, IGF2BP3, RUNX3 and
SFRP1 were associated with various diseases, including lung
cancer. Osteosarcoma has the potential to metastasize to the lungs
(3). As previously reported,
EGFR may be associated with osteosarcoma metastasis to the
lungs (37). In addition,
IGF2BP3 may promote cell invasiveness and tumor metastasis
of pancreatic cancer (38).
Therefore, the upregulation of EGFR and IGF2BP3
observed in the metastatic cells may indicate the metastatic
potential of osteosarcoma cells to the lung. Conversely, low
expression of RUNX3 may contribute to lymph node metastasis
and invasion in pancreatic carcinoma tissues (39). SFRP1 may inhibit
TGFβ-induced epithelial to mesenchymal transition phenotype and
thus inhibit metastasis (40).
Therefore, it is possible that the upregulation of RUNX3 and
SFRP1 may exhibit antitumor activity and have an opposite
effect on the metastasis of osteosarcoma to the lungs. Therefore,
it is possible that those four genes may be involved in the
metastasis of osteosarcoma to lung cancer. In addition, the
downregulated DEGs COL12A1, COL1A1, COL4A1 and
COL5A1 were associated with the extracellular collagen
biosynthesis pathway. In accordance with the enriched pathway,
those four genes were also enriched in biological processes
associated with extracellular matrix structure and organization.
Increased collagen degradation may promote the formation of primary
osteosarcoma tumors and metastasis to the lungs (41). The downregulation of the four genes
responsible for collagen synthesis may imply an impaired
extracellular matrix, which may contribute to tumor invasion in an
indirect manner.
A total of 10 TFs were screened from the DEGs,
including upregulated FOSL1, FOXA2 and PPARG,
and downregulated JUND. Based on the regulatory network of
TFs, SAT1 was targeted by TFs PPARG and JUND.
SAT1 deletion may lead to a significant reduction in BRCA1,
DNA repair associated expression (42), which may have a directly
suppressive effect on tumor metastasis-associated epithelial to
mesenchymal transition (43). It
is possible that upregulated SAT1 may be associated with
osteosarcoma metastasis. Activation of the upstream PPARG
may exert pro-tumorigenic effects on cancer progression and
metastasis in myeloid cells (44);
therefore, PPARG may be involved in the metastasis of
osteosarcoma through regulation of SAT1 expression. The
upstream JUND inhibition may suppress the migration, invasion and
metastasis of osteosarcoma (45).
Therefore, it is possible that upregulated PPARG and
downregulated JUND may exert opposite effects on
osteosarcoma metastasis via competitively regulating the expression
of SAT1. In addition, upregulation of the FOSL1
oncogene is involved in breast cancer migration and invasion
(46). FOXA2 may function
as a suppressor of lung cancer metastasis by inhibiting
TGF-β-induced epithelial to mesenchymal transition (47). The dysregulation of these 10 TFs
may suggest an involvement of TFs in metastasis.
The miRNA signature in osteosarcoma has been
extensively investigated using global microarray analyses of miRNAs
and mRNAs in osteosarcoma cell lines (48). Specific miRNAs targeting DEGs
involved in metastasis were identified in the present study.
Notably, two miRNAs with a high degree among these miRNAs were
detected. miR-21-5p had a regulatory effect on TPM1,
EGFR and TGFBR2, whereas miR-155-5p was able to
regulate E2F2. In a previous study, miR-155-5p was predicted
to be associated with the metastatic capacity of osteosarcoma
(49). It has previously been
reported that E2F2 has a key role in mediating tumor
metastasis of breast cancer (50).
Therefore, miR-155-5p may be involved in osteosarcoma metastasis
via targeting E2F2. In addition, miR-21-5p may contribute to
osteosarcoma metastasis via its target genes. EGFR has been
demonstrated to be associated with metastasis of osteosarcoma to
the lungs (37), whereas
TGFBR2 has been associated with metastasis of gastric cancer
cells (51), and suppressive
TPM1 may alter TGF-β tumor suppressor function and
thus promote metastasis of tumor cells (52). Therefore, it is possible that
miR-21-5p exerts a regulatory effect on metastasis via regulation
of the expression of its target genes.
In conclusion, upregulated PPP1R15A,
TGFBR2 and UCHL5, which are enriched in the
downregulation of TGF-β receptor signaling and TGF-β receptor
signaling activates SMADs pathways, may contribute to the
progression of osteosarcoma metastasis. In addition, the
downregulated DEGs BMP4, ID3 and SMAD6, which
are enriched in the TGF-β signaling pathway, may also be involved
in osteosarcoma metastasis. EGFR, IGF2BP3,
RUNX3 and SFRP1 were associated with metastasis to
lung cancer. Furthermore, 10 TFs screened from DEGs, and various
miRNAs (e.g. miR-21-5p), may be associated with metastasis via
their target genes. The full understanding of the complex
regulatory network of DEGs associated with metastatic osteosarcoma
may aid in improving the production of novel metastasis-targeted
therapeutic strategies.
References
|
1
|
Moore DD and Luu HH: Osteosarcoma. Cancer
Treat Res. 162:65–92. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Xing D, Qasem SA, Owusu K, Zhang K, Siegal
GP and Wei S: Changing prognostic factors in osteosarcoma: Analysis
of 381 cases from two institutions. Hum Pathol. 45:1688–1696. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Khanna C, Fan TM, Gorlick R, Helman LJ,
Kleinerman ES, Adamson PC, Houghton PJ, Tap WD, Welch DR, Steeg PS,
et al: Toward a drug development path that targets metastatic
progression in osteosarcoma. Clin Cancer Res. 20:4200–4209. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Dong Y, Liang G, Yuan B, Yang C, Gao R and
Zhou X: MALAT1 promotes the proliferation and metastasis of
osteosarcoma cells by activating the PI3K/Akt pathway. Tumour Biol.
36:1477–1486. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Brennecke P, Arlt MJ, Campanile C, Husmann
K, Gvozdenovic A, Apuzzo T, Thelen M, Born W and Fuchs B: CXCR4
antibody treatment suppresses metastatic spread to the lung of
intratibial human osteosarcoma xenografts in mice. Clin Exp
Metastasis. 31:339–349. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hou CH, Lin FL, Hou SM and Liu JF: Cyr61
promotes epithelial-mesenchymal transition and tumor metastasis of
osteosarcoma by Raf-1/MEK/ERK/Elk-1/TWIST-1 signaling pathway. Mol
Cancer. 13:2362014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhang Y, Zhang L, Zhang G, Li S, Duan J,
Cheng J, Ding G, Zhou C, Zhang J, Luo P, et al: Osteosarcoma
metastasis: Prospective role of ezrin. Tumour Biol. 35:5055–5059.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tu B, Peng ZX, Fan QM, Du L, Yan W and
Tang TT: Osteosarcoma cells promote the production of pro-tumor
cytokines in mesenchymal stem cells by inhibiting their osteogenic
differentiation through the TGF-β/Smad2/3 pathway. Exp Cell Res.
320:164–173. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Darnell JE Jr: Transcription factors as
targets for cancer therapy. Nat Rev Cancer. 2:740–749. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
van Kouwenhove M, Kedde M and Agami R:
MicroRNA regulation by RNA-binding proteins and its implications
for cancer. Nat Rev Cancer. 11:644–656. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
van der Deen M, Akech J, Lapointe D, Gupta
S, Young DW, Montecino MA, Galindo M, Lian JB, Stein JL, Stein GS
and van Wijnen AJ: Genomic promoter occupancy of runt-related
transcription factor RUNX2 in osteosarcoma cells identifies genes
involved in cell adhesion and motility. J Biol Chem. 287:4503–4517.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Liu H, Chen Y, Zhou F, Jie L, Pu L, Ju J,
Li F, Dai Z, Wang X and Zhou S: Sox9 regulates hyperexpression of
Wnt1 and Fzd1 in human osteosarcoma tissues and cells. Int J Clin
Exp Pathol. 7:4795–4805. 2014.PubMed/NCBI
|
|
13
|
Rubin EM, Guo Y, Tu K, Xie J, Zi X and
Hoang BH: Wnt inhibitory factor 1 decreases tumorigenesis and
metastasis in osteosarcoma. Mol Cancer Ther. 9:731–741. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhou G, Shi X, Zhang J, Wu S and Zhao J:
MicroRNAs in osteosarcoma: From biological players to clinical
contributors, a review. J Int Med Res. 41:1–12. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Osaki M, Takeshita F, Sugimoto Y, Kosaka
N, Yamamoto Y, Yoshioka Y, Kobayashi E, Yamada T, Kawai A, Inoue T,
et al: MicroRNA-143 regulates human osteosarcoma metastasis by
regulating matrix metalloprotease-13 expression. Mol Ther.
19:1123–1130. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Duan Z, Choy E, Harmon D, Liu X, Susa M,
Mankin H and Hornicek F: MicroRNA-199a-3p is downregulated in human
osteosarcoma and regulates cell proliferation and migration. Mol
Cancer Ther. 10:1337–1345. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yan K, Gao J, Yang T, Ma Q, Qiu X, Fan Q
and Ma B: MicroRNA-34a inhibits the proliferation and metastasis of
osteosarcoma cells both in vitro and in vivo. PLoS One.
7:e337782012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Barrett T and Edgar R: Gene expression
omnibus: Microarray data storage, submission, retrieval, and
analysis. Methods Enzymol. 411:352–369. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Altman NS: An introduction to kernel and
nearest-neighbor nonparametric regression. Am Stat. 46:175–185.
1992. View Article : Google Scholar
|
|
20
|
Hastie T, Tibshirani R, Narasimhan B and
Chu G: Impute: Imputation for microarray data. Bioconductor.
2016.
|
|
21
|
Bolstad B: preprocessCore: A collection of
pre-processing functions. R Package Version 1. 2013.
|
|
22
|
Amp TW: The Wadsworth & Brooks/Cole
mathematics series. Fourier Analysis & Its Applications.
|
|
23
|
Gentleman R, Carey V and Huber W:
Genefilter: Genefilter: Methods for filtering genes from microarray
experiments. R Package Version 1. 2007.
|
|
24
|
Smyth GK: Limma: Linear models for
microarray dataBioinformatics and computational biology solutions
using R and Bioconductor. Springer; pp. 397–420. 2005, View Article : Google Scholar
|
|
25
|
Ferreira JA and Zwinderman AH: On the
Benjamini-Hochberg method. Ann Stat. 34:1827–1849. 2006. View Article : Google Scholar
|
|
26
|
Chen YA, Tripathi LP and Mizuguchi K:
TargetMine, an integrated data warehouse for candidate gene
prioritisation and target discovery. PLoS One. 6:e178442011.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kohl M, Wiese S and Warscheid B:
Cytoscape: Software for visualization and analysis of biological
networksData Mining in Proteomics. Springer; pp. 291–303. 2011
|
|
28
|
Diao CY, Guo HB, Ouyang YR, Zhang HC, Liu
LH, Bu J, Wang ZH and Xiao T: Screening for metastatic osteosarcoma
biomarkers with a DNA microarray. Asian Pac J Cancer Prev.
15:1817–1822. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Xu S, Yang S, Sun G, Huang W and Zhang Y:
Transforming growth factor-beta polymorphisms and serum level in
the development of osteosarcoma. DNA Cell Biol. 33:802–806. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Nikitovic D, Zafiropoulos A, Katonis P,
Tsatsakis A, Theocharis AD, Karamanos NK and Tzanakakis GN:
Transforming growth factor-beta as a key molecule triggering the
expression of versican isoforms v0 and v1, hyaluronan synthase-2
and synthesis of hyaluronan in malignant osteosarcoma cells. IUBMB
Life. 58:47–53. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Lamora A, Talbot J, Bougras G, Amiaud J,
Leduc M, Chesneau J, Taurelle J, Stresing V, Le Deley MC, Heymann
MF, et al: Overexpression of smad7 blocks primary tumor growth and
lung metastasis development in osteosarcoma. Clin Cancer Res.
20:5097–5112. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Shi W, Sun C, He B, Xiong W, Shi X, Yao D
and Cao X: GADD34-PP1c recruited by Smad7 dephosphorylates TGFbeta
type I receptor. J Cell Biol. 164:291–300. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wicks SJ, Haros K, Maillard M, Song L,
Cohen RE, Dijke PT and Chantry A: The deubiquitinating enzyme UCH37
interacts with Smads and regulates TGF-beta signalling. Oncogene.
24:8080–8084. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Cao Y, Slaney CY, Bidwell BN, Parker BS,
Johnstone CN, Rautela J, Eckhardt BL and Anderson RL: BMP4 inhibits
breast cancer metastasis by blocking myeloid-derived suppressor
cell activity. Cancer Res. 74:5091–5102. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
DiVito KA, Simbulan-Rosenthal CM, Chen YS,
Trabosh VA and Rosenthal DS: Id2, Id3 and Id4 overcome a
Smad7-mediated block in tumorigenesis, generating TGF-β-independent
melanoma. Carcinogenesis. 35:951–958. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Jung SM, Lee JH, Park J, Oh YS, Lee SK,
Park JS, Lee YS, Kim JH, Lee JY, Bae YS, et al: Smad6 inhibits
non-canonical TGF-β1 signalling by recruiting the deubiquitinase
A20 to TRAF6. Nat Commun. 4:25622013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Selvarajah GT, Verheije MH, Kik M, Slob A,
Rottier PJ, Mol JA and Kirpensteijn J: Expression of epidermal
growth factor receptor in canine osteosarcoma: Association with
clinicopathological parameters and prognosis. Vet J. 193:412–419.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Taniuchi K, Furihata M, Hanazaki K, Saito
M and Saibara T: IGF2BP3-mediated translation in cell protrusions
promotes cell invasiveness and metastasis of pancreatic cancer.
Oncotarget. 5:6832–6845. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Xue LN, Bai FH, Wang XY, Lin M, Tan Y, Yao
XY and Xu KQ: Expression of RUNX3 gene in pancreatic adenocarcinoma
and its clinical significance. Genet Mol Res. 13:3940–3946. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Ren J, Wang R, Huang G, Song H, Chen Y and
Chen L: sFRP1 inhibits epithelial-mesenchymal transition in A549
human lung adenocarcinoma cell line. Cancer Biother Radiopharm.
28:565–571. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Husmann K, Arlt MJ, Muff R, Langsam B,
Bertz J, Born W and Fuchs B: Matrix Metalloproteinase 1 promotes
tumor formation and lung metastasis in an intratibial injection
osteosarcoma mouse model. Biochim Biophys Acta. 1832:347–354. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Brett-Morris A, Wright BM, Seo Y,
Pasupuleti V, Zhang J, Lu J, Spina R, Bar EE, Gujrati M, Schur R,
et al: The polyamine catabolic enzyme SAT1 modulates tumorigenesis
and radiation response in GBM. Cancer Res. 74:6925–6934. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Bai F, Chan HL, Scott A, Smith MD, Fan C,
Herschkowitz JI, Perou CM, Livingstone AS, Robbins DJ, Capobianco
AJ and Pei XH: BRCA1 suppresses epithelial-to-mesenchymal
transition and stem cell dedifferentiation during mammary and tumor
development. Cancer Res. 74:6161–6172. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Li H, Sorenson AL, Poczobutt J, Amin J,
Joyal T, Sullivan T, Crossno JT Jr, Weiser-Evans MC and Nemenoff
RA: Activation of PPARγ in myeloid cells promotes lung cancer
progression and metastasis. PLoS One. 6:e281332011. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Leaner VD, Chick JF, Donninger H, Linniola
I, Mendoza A, Khanna C and Birrer MJ: Inhibition of AP-1
transcriptional activity blocks the migration, invasion, and
experimental metastasis of murine osteosarcoma. Am J Pathol.
174:265–275. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Yang S, Li Y, Gao J, Zhang T, Li S, Luo A,
Chen H, Ding F, Wang X and Liu Z: MicroRNA-34 suppresses breast
cancer invasion and metastasis by directly targeting Fra-1.
Oncogene. 32:4294–4303. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Tang Y, Shu G, Yuan X, Jing N and Song J:
FOXA2 functions as a suppressor of tumor metastasis by inhibition
of epithelial-to-mesenchymal transition in human lung cancers. Cell
Res. 21:316–326. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Namlos HM, Meza-Zepeda LA, Barøy T,
Østensen IH, Kresse SH, Kuijjer ML, Serra M, Bürger H,
Cleton-Jansen AM and Myklebost O: Modulation of the osteosarcoma
expression phenotype by microRNAs. PLoS One. 7:e480862012.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Lauvrak SU, Munthe E, Kresse SH, Stratford
EW, Namløs HM, Meza-Zepeda LA and Myklebost O: Functional
characterisation of osteosarcoma cell lines and identification of
mRNAs and miRNAs associated with aggressive cancer phenotypes. Br J
Cancer. 109:2228–2236. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Hollern DP, Honeysett J, Cardiff RD and
Andrechek ER: The E2F transcription factors regulate tumor
development and metastasis in a mouse model of metastatic breast
cancer. Mol Cell Biol. 34:3229–3243. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Nadauld LD, Garcia S, Natsoulis G, Bell
JM, Miotke L, Hopmans ES, Xu H, Pai RK, Palm C, Regan JF, et al:
Metastatic tumor evolution and organoid modeling implicate TGFBR2
as a cancer driver in diffuse gastric cancer. Genome Biol.
15:4282014. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Varga AE, Stourman NV, Zheng Q, Safina AF,
Quan L, Li X, Sossey-Alaoui K and Bakin AV: Silencing of the
Tropomyosin-1 gene by DNA methylation alters tumor suppressor
function of TGF-beta. Oncogene. 24:5043–5052. 2005. View Article : Google Scholar : PubMed/NCBI
|