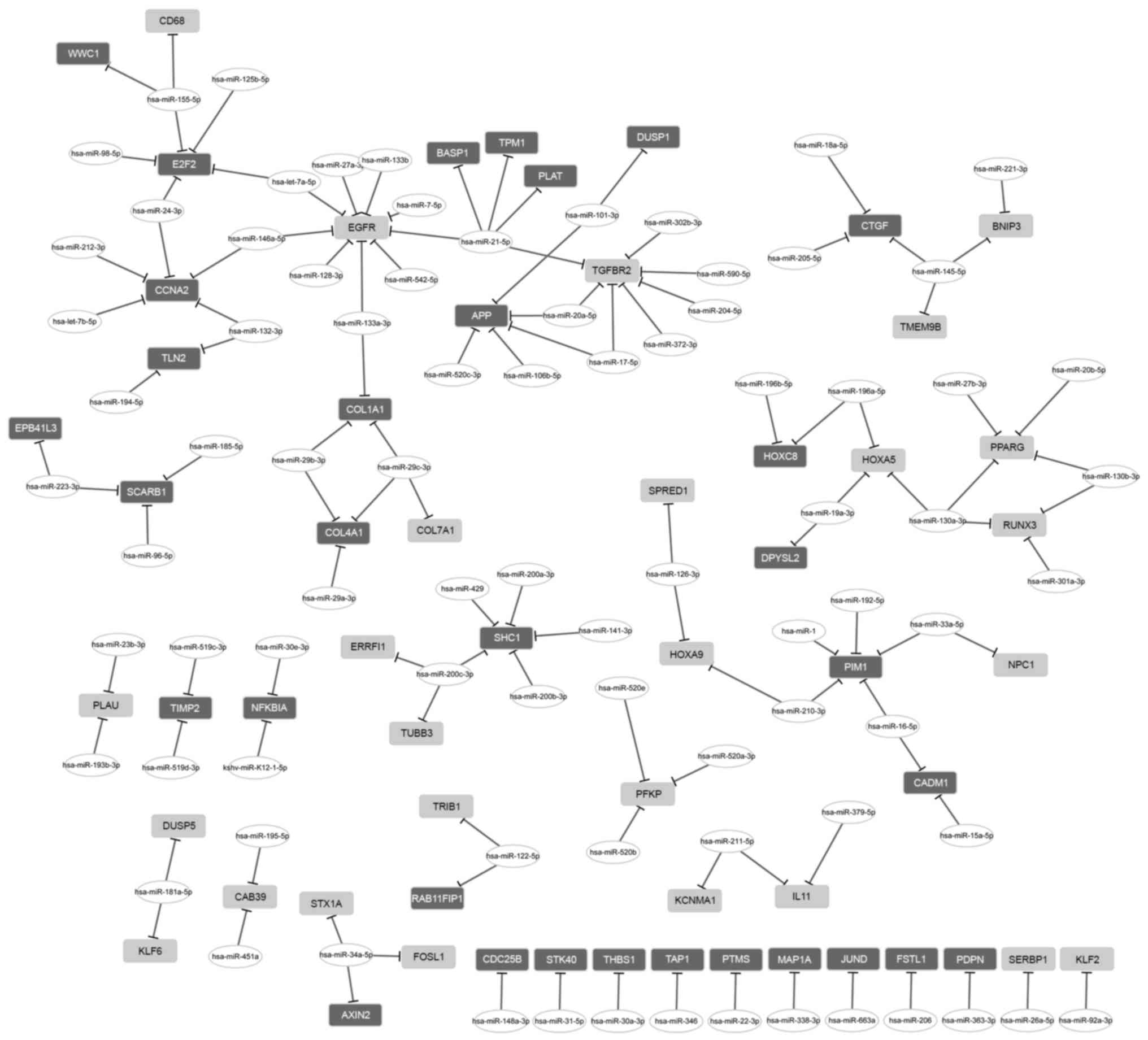
|
1
|
Moore DD and Luu HH: Osteosarcoma. Cancer
Treat Res. 162:65–92. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Xing D, Qasem SA, Owusu K, Zhang K, Siegal
GP and Wei S: Changing prognostic factors in osteosarcoma: Analysis
of 381 cases from two institutions. Hum Pathol. 45:1688–1696. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Khanna C, Fan TM, Gorlick R, Helman LJ,
Kleinerman ES, Adamson PC, Houghton PJ, Tap WD, Welch DR, Steeg PS,
et al: Toward a drug development path that targets metastatic
progression in osteosarcoma. Clin Cancer Res. 20:4200–4209. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Dong Y, Liang G, Yuan B, Yang C, Gao R and
Zhou X: MALAT1 promotes the proliferation and metastasis of
osteosarcoma cells by activating the PI3K/Akt pathway. Tumour Biol.
36:1477–1486. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Brennecke P, Arlt MJ, Campanile C, Husmann
K, Gvozdenovic A, Apuzzo T, Thelen M, Born W and Fuchs B: CXCR4
antibody treatment suppresses metastatic spread to the lung of
intratibial human osteosarcoma xenografts in mice. Clin Exp
Metastasis. 31:339–349. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hou CH, Lin FL, Hou SM and Liu JF: Cyr61
promotes epithelial-mesenchymal transition and tumor metastasis of
osteosarcoma by Raf-1/MEK/ERK/Elk-1/TWIST-1 signaling pathway. Mol
Cancer. 13:2362014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhang Y, Zhang L, Zhang G, Li S, Duan J,
Cheng J, Ding G, Zhou C, Zhang J, Luo P, et al: Osteosarcoma
metastasis: Prospective role of ezrin. Tumour Biol. 35:5055–5059.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tu B, Peng ZX, Fan QM, Du L, Yan W and
Tang TT: Osteosarcoma cells promote the production of pro-tumor
cytokines in mesenchymal stem cells by inhibiting their osteogenic
differentiation through the TGF-β/Smad2/3 pathway. Exp Cell Res.
320:164–173. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Darnell JE Jr: Transcription factors as
targets for cancer therapy. Nat Rev Cancer. 2:740–749. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
van Kouwenhove M, Kedde M and Agami R:
MicroRNA regulation by RNA-binding proteins and its implications
for cancer. Nat Rev Cancer. 11:644–656. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
van der Deen M, Akech J, Lapointe D, Gupta
S, Young DW, Montecino MA, Galindo M, Lian JB, Stein JL, Stein GS
and van Wijnen AJ: Genomic promoter occupancy of runt-related
transcription factor RUNX2 in osteosarcoma cells identifies genes
involved in cell adhesion and motility. J Biol Chem. 287:4503–4517.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Liu H, Chen Y, Zhou F, Jie L, Pu L, Ju J,
Li F, Dai Z, Wang X and Zhou S: Sox9 regulates hyperexpression of
Wnt1 and Fzd1 in human osteosarcoma tissues and cells. Int J Clin
Exp Pathol. 7:4795–4805. 2014.PubMed/NCBI
|
|
13
|
Rubin EM, Guo Y, Tu K, Xie J, Zi X and
Hoang BH: Wnt inhibitory factor 1 decreases tumorigenesis and
metastasis in osteosarcoma. Mol Cancer Ther. 9:731–741. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhou G, Shi X, Zhang J, Wu S and Zhao J:
MicroRNAs in osteosarcoma: From biological players to clinical
contributors, a review. J Int Med Res. 41:1–12. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Osaki M, Takeshita F, Sugimoto Y, Kosaka
N, Yamamoto Y, Yoshioka Y, Kobayashi E, Yamada T, Kawai A, Inoue T,
et al: MicroRNA-143 regulates human osteosarcoma metastasis by
regulating matrix metalloprotease-13 expression. Mol Ther.
19:1123–1130. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Duan Z, Choy E, Harmon D, Liu X, Susa M,
Mankin H and Hornicek F: MicroRNA-199a-3p is downregulated in human
osteosarcoma and regulates cell proliferation and migration. Mol
Cancer Ther. 10:1337–1345. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yan K, Gao J, Yang T, Ma Q, Qiu X, Fan Q
and Ma B: MicroRNA-34a inhibits the proliferation and metastasis of
osteosarcoma cells both in vitro and in vivo. PLoS One.
7:e337782012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Barrett T and Edgar R: Gene expression
omnibus: Microarray data storage, submission, retrieval, and
analysis. Methods Enzymol. 411:352–369. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Altman NS: An introduction to kernel and
nearest-neighbor nonparametric regression. Am Stat. 46:175–185.
1992. View Article : Google Scholar
|
|
20
|
Hastie T, Tibshirani R, Narasimhan B and
Chu G: Impute: Imputation for microarray data. Bioconductor.
2016.
|
|
21
|
Bolstad B: preprocessCore: A collection of
pre-processing functions. R Package Version 1. 2013.
|
|
22
|
Amp TW: The Wadsworth & Brooks/Cole
mathematics series. Fourier Analysis & Its Applications.
|
|
23
|
Gentleman R, Carey V and Huber W:
Genefilter: Genefilter: Methods for filtering genes from microarray
experiments. R Package Version 1. 2007.
|
|
24
|
Smyth GK: Limma: Linear models for
microarray dataBioinformatics and computational biology solutions
using R and Bioconductor. Springer; pp. 397–420. 2005, View Article : Google Scholar
|
|
25
|
Ferreira JA and Zwinderman AH: On the
Benjamini-Hochberg method. Ann Stat. 34:1827–1849. 2006. View Article : Google Scholar
|
|
26
|
Chen YA, Tripathi LP and Mizuguchi K:
TargetMine, an integrated data warehouse for candidate gene
prioritisation and target discovery. PLoS One. 6:e178442011.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kohl M, Wiese S and Warscheid B:
Cytoscape: Software for visualization and analysis of biological
networksData Mining in Proteomics. Springer; pp. 291–303. 2011
|
|
28
|
Diao CY, Guo HB, Ouyang YR, Zhang HC, Liu
LH, Bu J, Wang ZH and Xiao T: Screening for metastatic osteosarcoma
biomarkers with a DNA microarray. Asian Pac J Cancer Prev.
15:1817–1822. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Xu S, Yang S, Sun G, Huang W and Zhang Y:
Transforming growth factor-beta polymorphisms and serum level in
the development of osteosarcoma. DNA Cell Biol. 33:802–806. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Nikitovic D, Zafiropoulos A, Katonis P,
Tsatsakis A, Theocharis AD, Karamanos NK and Tzanakakis GN:
Transforming growth factor-beta as a key molecule triggering the
expression of versican isoforms v0 and v1, hyaluronan synthase-2
and synthesis of hyaluronan in malignant osteosarcoma cells. IUBMB
Life. 58:47–53. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Lamora A, Talbot J, Bougras G, Amiaud J,
Leduc M, Chesneau J, Taurelle J, Stresing V, Le Deley MC, Heymann
MF, et al: Overexpression of smad7 blocks primary tumor growth and
lung metastasis development in osteosarcoma. Clin Cancer Res.
20:5097–5112. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Shi W, Sun C, He B, Xiong W, Shi X, Yao D
and Cao X: GADD34-PP1c recruited by Smad7 dephosphorylates TGFbeta
type I receptor. J Cell Biol. 164:291–300. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wicks SJ, Haros K, Maillard M, Song L,
Cohen RE, Dijke PT and Chantry A: The deubiquitinating enzyme UCH37
interacts with Smads and regulates TGF-beta signalling. Oncogene.
24:8080–8084. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Cao Y, Slaney CY, Bidwell BN, Parker BS,
Johnstone CN, Rautela J, Eckhardt BL and Anderson RL: BMP4 inhibits
breast cancer metastasis by blocking myeloid-derived suppressor
cell activity. Cancer Res. 74:5091–5102. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
DiVito KA, Simbulan-Rosenthal CM, Chen YS,
Trabosh VA and Rosenthal DS: Id2, Id3 and Id4 overcome a
Smad7-mediated block in tumorigenesis, generating TGF-β-independent
melanoma. Carcinogenesis. 35:951–958. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Jung SM, Lee JH, Park J, Oh YS, Lee SK,
Park JS, Lee YS, Kim JH, Lee JY, Bae YS, et al: Smad6 inhibits
non-canonical TGF-β1 signalling by recruiting the deubiquitinase
A20 to TRAF6. Nat Commun. 4:25622013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Selvarajah GT, Verheije MH, Kik M, Slob A,
Rottier PJ, Mol JA and Kirpensteijn J: Expression of epidermal
growth factor receptor in canine osteosarcoma: Association with
clinicopathological parameters and prognosis. Vet J. 193:412–419.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Taniuchi K, Furihata M, Hanazaki K, Saito
M and Saibara T: IGF2BP3-mediated translation in cell protrusions
promotes cell invasiveness and metastasis of pancreatic cancer.
Oncotarget. 5:6832–6845. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Xue LN, Bai FH, Wang XY, Lin M, Tan Y, Yao
XY and Xu KQ: Expression of RUNX3 gene in pancreatic adenocarcinoma
and its clinical significance. Genet Mol Res. 13:3940–3946. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Ren J, Wang R, Huang G, Song H, Chen Y and
Chen L: sFRP1 inhibits epithelial-mesenchymal transition in A549
human lung adenocarcinoma cell line. Cancer Biother Radiopharm.
28:565–571. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Husmann K, Arlt MJ, Muff R, Langsam B,
Bertz J, Born W and Fuchs B: Matrix Metalloproteinase 1 promotes
tumor formation and lung metastasis in an intratibial injection
osteosarcoma mouse model. Biochim Biophys Acta. 1832:347–354. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Brett-Morris A, Wright BM, Seo Y,
Pasupuleti V, Zhang J, Lu J, Spina R, Bar EE, Gujrati M, Schur R,
et al: The polyamine catabolic enzyme SAT1 modulates tumorigenesis
and radiation response in GBM. Cancer Res. 74:6925–6934. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Bai F, Chan HL, Scott A, Smith MD, Fan C,
Herschkowitz JI, Perou CM, Livingstone AS, Robbins DJ, Capobianco
AJ and Pei XH: BRCA1 suppresses epithelial-to-mesenchymal
transition and stem cell dedifferentiation during mammary and tumor
development. Cancer Res. 74:6161–6172. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Li H, Sorenson AL, Poczobutt J, Amin J,
Joyal T, Sullivan T, Crossno JT Jr, Weiser-Evans MC and Nemenoff
RA: Activation of PPARγ in myeloid cells promotes lung cancer
progression and metastasis. PLoS One. 6:e281332011. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Leaner VD, Chick JF, Donninger H, Linniola
I, Mendoza A, Khanna C and Birrer MJ: Inhibition of AP-1
transcriptional activity blocks the migration, invasion, and
experimental metastasis of murine osteosarcoma. Am J Pathol.
174:265–275. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Yang S, Li Y, Gao J, Zhang T, Li S, Luo A,
Chen H, Ding F, Wang X and Liu Z: MicroRNA-34 suppresses breast
cancer invasion and metastasis by directly targeting Fra-1.
Oncogene. 32:4294–4303. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Tang Y, Shu G, Yuan X, Jing N and Song J:
FOXA2 functions as a suppressor of tumor metastasis by inhibition
of epithelial-to-mesenchymal transition in human lung cancers. Cell
Res. 21:316–326. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Namlos HM, Meza-Zepeda LA, Barøy T,
Østensen IH, Kresse SH, Kuijjer ML, Serra M, Bürger H,
Cleton-Jansen AM and Myklebost O: Modulation of the osteosarcoma
expression phenotype by microRNAs. PLoS One. 7:e480862012.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Lauvrak SU, Munthe E, Kresse SH, Stratford
EW, Namløs HM, Meza-Zepeda LA and Myklebost O: Functional
characterisation of osteosarcoma cell lines and identification of
mRNAs and miRNAs associated with aggressive cancer phenotypes. Br J
Cancer. 109:2228–2236. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Hollern DP, Honeysett J, Cardiff RD and
Andrechek ER: The E2F transcription factors regulate tumor
development and metastasis in a mouse model of metastatic breast
cancer. Mol Cell Biol. 34:3229–3243. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Nadauld LD, Garcia S, Natsoulis G, Bell
JM, Miotke L, Hopmans ES, Xu H, Pai RK, Palm C, Regan JF, et al:
Metastatic tumor evolution and organoid modeling implicate TGFBR2
as a cancer driver in diffuse gastric cancer. Genome Biol.
15:4282014. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Varga AE, Stourman NV, Zheng Q, Safina AF,
Quan L, Li X, Sossey-Alaoui K and Bakin AV: Silencing of the
Tropomyosin-1 gene by DNA methylation alters tumor suppressor
function of TGF-beta. Oncogene. 24:5043–5052. 2005. View Article : Google Scholar : PubMed/NCBI
|